Received: January 9, 2014
Accepted: May 9, 2014
Meditterr J Hematol Infect Dis 2014, 6(1): e2014038, DOI 10.4084/MJHID.2014.038
This article is available on PDF format at:

Ghaleb Elyamany1,2, Mohammed Awad2, Omar Alsuhaibani2, Kamal Fadalla3, Omer Al Sharif4, Mohammad Al Shahrani4, FahadAlabbas4 and Abdulaziz Al-Abulaaly3
1
Department of Hematology and Blood Bank, Theodor Bilharz Research
Institute.
2 Dept. of Central Military Laboratory, Prince
Sultan Military Medical City, Riyadh, Saudi Arabia.
3 Dept. of Adult Clinical Hematology and Stem
cell Therapy, Prince Sultan Military Medical City, Riyadh, Saudi Arabia.
4 Dept. of Pediatric Hematology/Oncology, Prince
Sultan Military Medical City, Riyadh, Saudi Arabia.

|
This
is an Open Access article distributed
under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract The
fms-like tyrosine kinase 3 (FLT3) gene is a member of the class III
receptor tyrosine kinase family. Mutations of FLT3 were first described
in 1997and account for the most frequent molecular mutations in acute
myeloid leukemia.
Currently, there is no published data on FLT3 mutations in Saudi acute lymphoblastic leukemia (ALL) patients. In this retrospective study, we have examined a cohort of 77 ALL patients to determine the prevalence of FLT3 mutations and the possible prognostic relevance of these mutations in ALL patients. Correlations to other biologic factors such as karyotype, molecular mutations, and leukocyte count were also considered. FLT3 internal tandem duplication (ITD) mutations and point mutation in tyrosine kinase domain (D835) were analyzed in ALL patients, at diagnosis, by polymerase chain reaction (PCR). Two cases (2.6%, 2/77) were positive for FLT3 mutations; one was found to have FLT3/ITD and the other FLT3/D835. Our findings suggest that FLT3 mutations are not common in Saudi ALL and do not affect clinical outcome. |
Introduction
The
human fms-like tyrosine kinase 3 (FLT3) gene is located on chromosome
13q12 and encompasses 24 exons. It encodes a membrane-bound
glycosylated protein of 993 amino acids with a molecular weight of
158-160kDa, as well as a non-glycosylated isoform of 130-143 kDa that
is not associated with the plasma membrane.[1]
The structure of FLT3 is shown in the figure 1.[2]
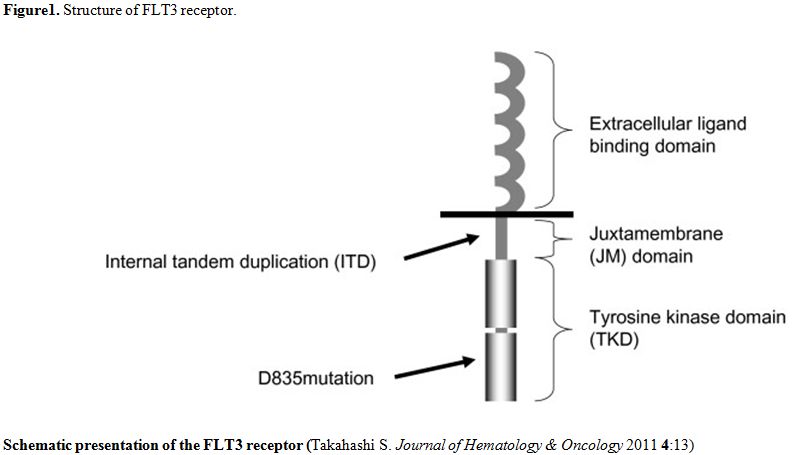 | Figure 1. Structure of FLT3 receptor. |
Genomic
aberrations of FLT3, including internal tandem duplication (ITD) and
point mutations, have been demonstrated in approximately 25–35% of
adults with acute myeloid leukemia (AML).[3-7]
ITD of
the FLT3 gene is common in AML and is associated with a bad prognosis
and poor response to chemotherapy. Single base mutations at the FLT3
tyrosine kinase domain (TKD), which frequently involves aspartic acid
835 of the kinase domain (D835), leads to a gain of function; however,
due to its rarity, its prognostic significance is not well defined.[8]
FLT3 is rarely mutated in leukemic lymphoblasts in adult and pediatric
ALL;[3,4,9,10]
however, FLT3 mutations are relatively common among the cytogenetic
subgroups of hyperdiploidy and mixed-lineage leukemia (MLL)
translocation.[11]
Recent studies have indicated a low overall frequency in childhood ALL
(in the 1-8% range) while consistently demonstrating a higher incidence
among those with MLL gene rearrangement and high hyperdiploidy.[13-17] In adult ALL, FLT3 mutations are
even rarer.[18]
While there have been several studies[19-27]
describing activating mutations of the FLT3 gene in AML, there has been
little work on these mutations in ALL.
In this study, we analyzed the prevalence of the two types of FLT3
activating mutations in 77 patients with ALL and its prognostic
significance. No data currently exist regarding FLT3 mutations in Saudi
ALL patients and this study is the first one conducted in Saudi Arabia
describing FLT3 mutations in ALL patients.
Introduction
Material and Methods
Study Group: A retrospective review of both adult and pediatric (ages 1 to 15) cases of ALL was performed. Data was obtained from the files of the Department of Hematopathology, Prince Sultan Military Medical City, Saudi Arabia from 2005 to 2013. Leukemia samples were obtained from either bone marrow (BM) or peripheral blood (PB), at diagnosis, from patients with ALL (70 BM samples and 7 PB samples). The peripheral blood samples all had more than 15% blasts at diagnosis. Five samples obtained from normal bone marrow healthy donors were screened for FLT3 mutations as a reference group. Among the 77 patients, with an established diagnosis by cell morphology and flow cytometric immunophenotyping, 48 were pediatric (62.3%), 29 were adult (37.7%), in total, 45 of the patients were male (58.4%) and 32 female (41.6%) Table 1.
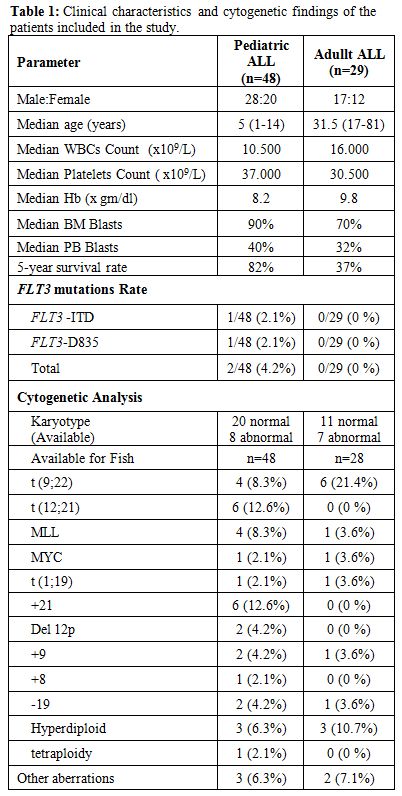 | Table 1. Clinical characteristics and cytogenetic findings of the patients included in the study. |
Samples
were evaluated in addition to cytomorphology, and multiparameter flow
cytometry, by cytogenetics, fluorescence in situ hybridization (FISH),
and molecular genetics in parallel.
Pediatric patients were treated according to the UKALL 2003
chemotherapy protocol. Initially, eligible pediatric patients were
stratified into three risk groups, standard risk (22 patients),
intermediate risk (14 patients) and high risk (12 patients) based on
age, WBC at presentation, immunophenotype and cytogenetic
abnormalities.
Treatment of adult ALL patients was divided into two age groups.
Patients at 20 years of age or less were treated according to the
Dana-Farber Cancer Institute All Consortium Protocol 00-01[28] and patients over 20 years of age
were treated with Hyper-CVAD chemotherapy.[29]
At first, risk groups at diagnosis were categorized into a standard
risk (15 patients) and high risk (14 patients) to determine the
intensity of therapy. This study was approved by the Research and
Ethics Committee at this institution.
Morphologic Analysis: For each case in this study, Wright-Giemsa-stained peripheral blood and bone marrow aspirate smears were reviewed. Aspirate clot and biopsy specimens were fixed in formalin, routinely processed and the histologic sections were stained with Hematoxylin and Eosin and reviewed.
Flow CytometricImmunophenotypic Methods: All samples were assessed by multicolor flow cytometry using a large panel of antibodies, including CD2, cytoplasmic and surface CD3, CD4,CD5, CD7, CD8, CD10, CD19,CD20, CD22, cytoplasmic CD79a, CD13, CD15, CD33, CD11c, CD14, CD64, CD38, CD34, CD117, cytoplasmic terminal deoxynucleotidyltransferase (TdT), and myeloperoxidase (MPO). Antigens were scored as positive using a cutoff of 20% or more leukemic blasts staining brighter than an isotype-matched negative control.
Molecular Methods:
Analysis of FLT3-ITD mutation. DNA was extracted using a QIAamp DNA
Kit (Qiagen) according to the manufacturer’s recommendations.PCR amplification was composed of 200ng of DNA, 50mM KCL, 10mM
Tris-HCL, pH8.3, 1.5mM MgCL2, 0.001%(wt/vol) gelatin, 200 µM dNTPs,
0.4µM of each primer (5’-GCAATTTAGGTATGAAAGCCAGC-3’ and
5’-CTTTCAGCATTTTGACGGCAACC-3’), and 1U of gold Taq polymerase, in a
volume of 50µl.[23]
The PCR consisted of an initial incubation step at 95°C for 10 minutes
followed by 35 cycles at 94°C for 30 seconds, 57°C for 60 seconds, and
72°C for 90 seconds. The final extension step was at 72°C for 10
minutes on a GeneAmp PCR system 9700(Applied Biosystems). PCR products
were analyzed on standard 3% agarose gels. Normal amplification
generates a 330bp product; whereas, FLT3 ITD mutations (FLT3/ITD+) show
longer PCR products.
Analysis of the FLT3- D835 mutation. PCR amplification was set up as above using specific primers[4] for exon 20 (5’-CCGCCAGGAACGTGCTTG-3’ and 5’-GCAGCCTCACATTGCCCC-3’). PCR product was digested with EcoRV (Promega), at 37°C for 2h. The digestion products were separated on a 3% agarose gel, and incomplete digestion indicated the presence of mutant.
Statistical
analysis:
The Kaplan-Meier technique was used to analyze the probability of
overall survival (OS). OS was calculated from time of diagnosis to
death. Continuous variables, such as white blood cell count and
hemoglobin, were compared using the Kruskal-Wallis test. Differences
between means were considered as significant at P < 0.05.
Complete remission (CR) was defined by less than 5% blast cells in a
normocellular marrow and peripheral blood neutrophil count equal to or
greater than 1.5 x 109/liter
with a platelet count of more than 100 x 109/liter.
Normalization of cytogenetic abnormalities was not a prerequisite for
CR.
Results
Clinical characteristics at presentation and cytogenetic analysis of
the patients in the studied group are summarized in table 1.
In total, the patient age range was 1 to 81 years with a median of 15
years. Pediatric patients were defined as less than 15 years of age. Of
the 77 patients, 48 were pediatric (62.3%), all with de novo ALL (42
cases B-ALL and 6 cases T-ALL). 29 were adult patients (37.7%) in which
there were 27 cases of de novo ALL (22 cases B-ALL and 5 cases T-ALL)
and 2 cases of CML transforming to ALL. Two cases from the pediatric
group were diagnosed as biphenotypic leukemia. In total, 45 of the
patients were male (58.4%) and 32 female (41.6%).
Two of the 77 ALL patients examined showed FLT3 mutations (2/77) with
an overall prevalence of 2.6%. Positive FLT3 mutation patients were
both pediatric, one male and one female. One was found to have FLT3/ITD
and the other was positive for FLT3/D835 (Figure 2).
None of the adult ALL patients was positive for FLT3 mutations. None of
the T-ALL (T-ALL - 11/77) and CML patients transforming to ALL showed
FLT3/ITD or D835 mutations. None of the patients had a combination of
FLT3/ITD and D835 mutation in the FLT3 gene.
The FLT3/ITD
positive patient (male, 14 years old) showed a WBC of 91 x 109/liter
with 70% of the peripheral blasts. The bone marrow (BM) blasts were 95%
and showed a good response to chemotherapy resulting in CR. The
FLT3/D835 positive patient (female, 4 years old) showed a WBC of 4.6 x
109/liter
with 32% of peripheral
blasts. The bone marrow (BM) blasts were 95%, and continuous complete
remission (CCR) was achieved after chemotherapy.
Cytogenetic and molecular studies revealed that the FLT3/D835 positive
patient was hyperdiploidy and reported as
54,XX,+2,+4,+8,+14,+16,+20,21,+21.nuc ish (ABL1,BCR) x2[100]/
(ETV6,RUNX1)x3[95/100]/(5'MLL,3'MLL,5'MLL con 3'MLL)x2[100]
/(5'MYC.3'MYC,5'MY con 3'MYC)x3 [95/100]/ (TCF3,PBX1)x2[100]. An extra
copy of chromosome 2, 4, 8, 12, 14, 16, 20 and 21 was detected in 95%
of the studied cells. Cytogenetic analysis could not be performed due
to an insufficient number of metaphases.
Discussion
FLT3 gene mutations, particularly ITD in AML, are well established as the most frequent somatic alterations in AML. A poor prognosis is associated with FTL3 gene mutations in AML and they are found in approximately 5-15% of children and 25-35% of adults with AML.[4,7,19,20,23,31] FLT3 mutations are also found in adult and pediatric ALL, but are much rarer than in AML.[3,4,9,10] FLT3 is overexpressed at the level of RNA and protein in most B lineage and acute myeloid leukemias. It is also overexpressed in a smaller subset of T-ALL and chronic myeloid leukemias (CML) in blast crisis.[32]
This study is the first to report FLT3 mutations on ALL patients in Saudi Arabia. The overall rate of patients with ALL and FLT3 mutations, in this study, was 2.6% which is comparable with previously published reports.[4,14,33,34,35]
The incidence of FLT3 mutations in pediatric ALL of this study was 4.2% (2/48). The results are within the range of similar studies conducted in different geographic regions (see table 2).
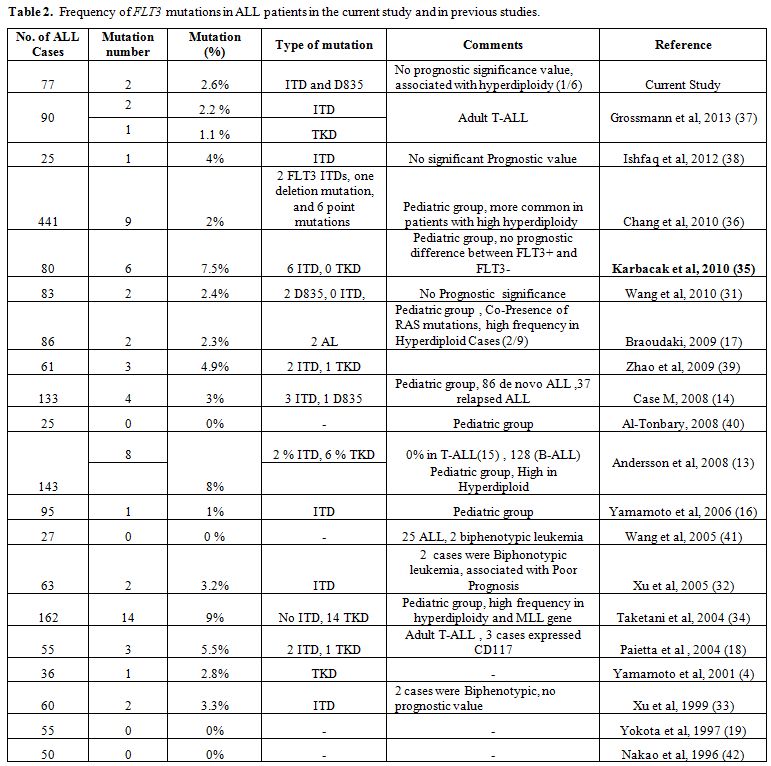 | Table 2. Frequency of FLT3 mutations in ALL patients in the current study and in previous studies. |
The
incidence of FLT3 mutations in pediatric leukemia is of particular
interest due to the several promising FLT3 inhibitors currently under
development.[45]
The frequency of both ITD and D835 mutations among adult ALL patients
in this study was 0% (0/29); other studies have reported similar
results;[19,43,44]
however, a few studies have reported FLT3 mutations among adult ALL
patients at a very low frequency.[18]
As this is a rare occurrence, the limited sample size of this study is
the most likely cause of the difference. Similarly, neither FLT3/ITD
nor FLT3/D835 mutations were evident in any case of CML transforming to
ALL or in T-All (0/11) which is consistent with the study by Andersson
et al. 2008 (0% in 15 T-ALL patients).[13]
Some
larger studies have reported a low frequency of FLT3/ITD and/or
FLT3/D835 mutations ranging from 3.3% to 5.5% among T-ALL patients.[18,39,46]
In this study, one patient (male, 14 years old) was found to have the
FLT3/ITD mutation. He showed leukocytosis and a high blast cell count
in PB and BM. This patient exhibited a good response to chemotherapy
and achieved complete remission, which is in-line with results of
previous studies.[35,37,39]
This patient was also positive for myeloid antigen expression and was
diagnosed as acute mixed-lineage leukemia (biphenotypic leukemia). This
observation was reported in other studies[17,34,35]
which indicates a higher frequency among the biphenotypic group.
Cytogenetic analysis could not be performed due to the low number of
metaphases.
The other FLT3 mutation patient in this study (female, 4 years old),
was found to the FLT3/D835 mutation with no leukocytosis; however, she
did show a high blast count in PB and BM. Cytogenetic and molecular
studies revealed no association with balanced translocations or MLL
gene rearrangement; however, hyperdiploidy (extra copy of chromosome 2,
4, 8, 12, 14, 16, 20 and 21) was detected in 95% of the studied cells.
This finding represents a 16.7% incidence of hyperdiploidy in ALL (1/6)
cases. Our results reinforced previous observations, Armstrong et al.
2004,[12] in that the presence of
the FLT3 mutation
in hyperdiploid ALL did not affect the clinical outcome as the patient
responded well to chemotherapy and achieved continuous complete
remission (CCR).
In both cases, molecular studies using polymerase chain reaction (PCR)
were negative for Philadelphia chromosome [t(9;22)(q34.1;q11.2)].
The only notable difference between this study and previous studies[11,12,36]
is the lack of association between the high frequency of FLT3 mutation
and MLL gene rearrangement. The small sample size (5 ALL patients) is
the most likely cause of the difference.
Neither ITD’s or D835 mutations were detected in the healthy donors of
this study, and this is consistent with previously reported data.[4,47]
Conclusion
FLT3 mutations exist in a small proportion of Saudi ALL patients.
Regarding clinical outcomes, there was no prognostic significance in
ALL patients with or without FLT3 mutations. The observation of high
frequency in hyperdiploid ALL is in agreement with similar studies from
other geographical regions; however, there is a lack of association
between MLL gene rearrangement and FLT3 mutations among ALL patient
that may be due to the limited sample size. Further studies are needed
to confirm and establish these results.
Acknowledgment
We thank Jon Johnston, Principal Clinical Scientist from Histopathology lab for editing this research article. We thank all the Hematology, Cytogenetic and Molecular Laboratories Staffs, especially Mr. Mohamed Asiri and Dalal Alkhammash from Hematology Lab for data collection and Ms Nadia Halawani from Molecular Lab for the great support in Molecular work of this study. We also thank Ms Khawla Al-Fayez from Cytogenetic Lab for excellent support and data collection in cytogenetic aspect of this study.
Materials and Methods
Umbilical
cord blood collection.
Between August 2010 and July 2011, 177 mothers delivering their babies
at Al-Isra'a hospital, Amman, Jordan, were approached to participate in
this prospective study. One hundred and twenty-four mothers (70%)
agreed to participate and signed a consent form. The UCB was collected
exclusively from term (gestation period 37-42 weeks) single-birth
babies born through normal vaginal delivery. Cord blood was collected
after the baby was delivered but before the delivery of the placenta. A
regular blood-donor set was used for UCB collection containing 28 ml
citrate phosphate dextrose-adenine (CPD-A) anticoagulant. The
collection was performed by the obstetrician delivering the baby and
not by a trained technician. The umbilical cord (UC) was sterilized
with povidone iodine in a unidirectional move, and 16-gauge needle of
the prepared blood-donor set was inserted into the umbilical vein.
Blood was allowed to flow by gravity, and the needle was removed when
blood flow ceased as has been previously described.[6,10]
The study design and UCB collection procedure was approved by the
Hashemite University, and Al-Isra'a general hospital Institute Review
Boards.
Evaluation
of umbilical cord blood parameters.
For the current study, UCB units were deemed unacceptable if the total
volume collected was less than 30 ml and/or if the unit was delivered
for analysis past 24 hours of collection. The UCB units were processed
and analyzed in the biology laboratory at the Hashemite University,
Jordan. The UCB was incubated with FITC-conjugated anti-CD45
fluorescein (MACS, Germany) and PE-conjugated anti-CD34 PE (MACS,
Germany) for 30 min at room temperature in the dark. After incubation,
RBCs were lysed with the lysis solution (Coulter, France) and then
washed twice with 10% bovine serum albumin (BSA) in phosphate buffer
saline (PBS). For each tube, 20,000 live events were counted in a
flowcytometer counter (Partec, Germany). CD34+
cells were selected based on their forward- and 90o-scatter
properties and dim CD45 expression.[13]
The clonogenic assay (CFU-GM assay) was performed as described
previously.[14] Briefly,
mononucleated cells (MNCs) were cultured at 1.0 X 105/ml
in RBMI-1640 medium (MACS, Germany) containing 0.8% methylcellulose
(Sigma Aldrich, USA), 20% fetal bovine serum (FBS; Lonza, Belgium), 450
µg/ml human transferrin, 10 ng/ml GM-CSF, 10 ng/ml IL-3 (Stem Cell
Technologies), and 1% BSA. Cells were incubated at 37oC
in a humidified atmosphere of 5% CO2
for 14 days. Colonies (clusters containing at least 50 cells) were
counted using an inverted microscope (Leica, Germany). Viability was
determined using trypan blue dye exclusion method, where the non-viable
cells stain deep blue.
Maternal
and neonatal data collection.
Data regarding maternal age, the number of previous pregnancies and
live births were collected from the medical files. Neonatal data such
as the weight of the baby and the placenta, baby’s gender, and UC
length were collected from the obstetric staff clinical notes at
Al-Isra'a general hospital. A standard questionnaire was prepared and
used for data collection.
Statistical
analysis.
Statistical analysis was carried out using STATISTICA 7 analysis
program (StatSoft Inc., OK, USA). Results were expressed as mean ±
standard deviation (SD). One-way analysis of variance (ANOVA) was used
to test for a significant difference between mean values of all.
Spearman's correlation was used to assess the association between the
different variables. A p value of ≤ 0.05 was considered statistically
significant.
Results
Characteristics
of the study population.
A total of 177 prospective mothers were approached to participate in
the current study, 53 (30%) of them refused to participate due to
cultural and/or lack of knowledge regarding benefits of UCB
and
safety of the collection procedure. 124 units were prospectively
collected for this study. In 17 (13.7%) UCB units the net volume of
cord blood was less than 30 ml, in 23 (18.5%) units some maternal
and/or neonatal data were missing, and in 9 (7.3%) units the samples
were not delivered for the laboratory within 24 hour of collection. A
total of 75 UCB units (60.5% of the total collected units) were
included and analyzed in this study. The characteristics of the
donating mothers and babies are shown in Table 1.
The mean maternal age was 28 years (range 19-43). Thirty three percent
of the donating mothers were delivering their first babies, 21.3%
second, 20% third and 25.3% fourth or more. Fifty one percent of the
delivered babies were males, and 49% were females. The mean weight of
the delivered babies was 3178 gm (range 220-4160), and mean placenta
weight was 526 gm (range 400-655). Seven percent of the donating
mothers reported that they were current smokers.
 |
Table 1. Maternal and neonatal characteristics |
Analysis
of umbilical cord blood samples.
Cord blood cell counts were analyzed within 24 hours of collection. The
mean volume of UCB collected (not including the 28 ml of anticoagulant)
was 68.9 ml (range 40-115 ml). The mean viability was 94.9% (range
80-99%), the mean total nucleated cell (TNC) count was 6.5 x 108 (range
1-32), with 10.6% have TNC of more than 1 x 109
and 4% of more than 1.2 x 109.
The mean total mononuclear cell count (MNC) was 3.4 x 108 (range
0.5-14.9), the mean total CD34+
cell count was 3.8 x 106
(range 0.2-11.8), and the mean total CFU-GM was 9.9 x105 (range
2-25). The UCB unit’s data are summarized in Table 2.
The results of the univariate analysis correlation are presented in Table 3. The volume
of UCB units collected was positively correlated with TNC (p=0.008),
cell viability (p=0.001), MNC (p=0.018), CD34+
cell count (p=0.034) and with the umbilical cord length (p=0.011).
There was also a trend towards obtaining higher UCB volume from mothers
with increasing number of prior live births (p=0.086). Our results
showed that higher TNC is correlated with MNC (p=0.001), CD34+
cell count (p=0.009), and increased viability (p=0.001). Finally, our
study demonstrated an inverse correlation between CFU-GM concentration
and the gestation duration (P = 0.038). There was no significant effect
of gestational age on TNC or CD34+
cell count of the collected UCB.
 |
Table 2. Analysis of umbilical cord blood units (No 75) |
 |
Table 3. Univariate analysis for correlation |
Discussion
This
is the first study to show that collection of cord blood is
feasible and can result in adequate TNC collection and viability in a
developing country in the Middle East. Of the 124 women enrolled in the
study, the umbilical cord blood of only 17 (13.7%) did not contain
adequate volume of blood despite the fact that untrained technicians
were present at the delivery to collect the UCB. An additional 25.8% of
the UCB units had to be excluded either because some data were missing,
or because the UCB did not reach the lab in the required time for
processing. Among the 75 units that met the predefined eligibility
criteria, the volume, nucleated cell dose and CD34 count was similar to
what has been previously published.[5,7,8,10,14]
Previous studies concluded that UCB yield of TNC, CD34+
cells, and CFU-GM is influenced not only by neonatal and maternal
factors but also by ethnicity of the parents.[7,15]
In this study, a total of 75 cord blood samples from Jordanian neonates
were analyzed in order to investigate any neonatal and maternal factors
that might influence UCB unit in terms of TNC and CD34+
cell content, and CFU-GM yields. In the current study, the average age
of the donor mothers included was 28 years and both the TNC and CD34+5,8-10]
We found no significant
correlation between birth order and UCB volume, TNC, and CD34+
levels. Our findings are different from prior studies which showed that
women with few previous live births produced UCB units with higher TNC
yields.[9,10] Cell yield was not
influenced by maternal age. While the majority of
published data showed similar observation, the study by Nakagawa et al.
which analyzed 572 UCB units, showed that increasing mother’s age was
associated with lower TNC cell yield.
The volume of UCB collected was not influenced by any of the maternal
or neonatal factors except the umbilical cord length, as longer cords
were associated with higher volume collected. This correlation has only
been observed in one prior study from Japan.[8]
We also found an inverse correlation between the CFU-GM concentration
and gestational age, which indicates that there is a loss of
hematopoietic potential with a longer gestational age. In a study by
Ballen et al. of 1269 UCB units, there was an 11% decrease in CFU-GM
with each additional week of gestation.[10]
Since TNC and CD34+
cell counts are the most important predictors of the outcome following
cord blood transplant, collecting units with a larger volume is
desired. In our current study, the only positive predictor of improved
cell count is the volume of UCB collected.
The chance of finding a matched related donor in Jordan is
significantly higher than what has been observed in other countries
(65% versus 25%) due to more homogeneous ethnic group in the region.[16]
Approximately 10-16% of UBC units collected in the international cord
blood banks have TNC of more than 1.2 x 109.
In Our study, 10.6% of UCB units collected have TNC of more than 1 x 109 and 4% of
more than 1.2 x 109.
Although our cell dose was slightly lower than what has been reported
by established cord blood banks, this is dependent on the experience of
the collection staff which always improves with time.
We believe that establishing a cord blood bank in Jordan will further
increase the possibility of identifying donors for patients who lack
related donor options. Taking into consideration the geographical and
cultural similarities between Jordan and its neighboring Arabic
countries, a cord blood bank in Jordan will help patients throughout
the region. Additional training and better logistical support are
needed to collect UCB units in order to decrease the percentage of
unacceptable units collected. We need also more efforts towards
education of the parents about the benefits and safety of UCB
collection, as only 70% of the approached mothers agreed to participate
in this study. A proper cost-effective analysis should be carried out
before establishing national cord blood banks in countries with limited
resources.
In conclusion, we have found that collection of cord blood units in
Jordan is feasible and can result in similar cell content compared to
other developed countries. Efforts toward establishing public cord
blood banks in our area are warranted.
Acknowledgments
We
are grateful for the effort of the attending physicians, residents
and nursing staff of Obstetrics and Gynecology department at Al-Isra'a
Hospital. We gratefully acknowledge the Deanship of Research and
Graduate Studies, The Hashemite University, Jordan for financial
support.
Author contribution: Study design (AAH, RMB, LHT, AZE), study analysis
and interpretation of the data (AAH, RMB, LHT, HF, AZE). All authors
contributed to the writing of the manuscript and approval of the final
version.
References















































[TOP]