Received: August 11, 2015
Accepted: November 11, 2015
Mediterr J Hematol Infect Dis 2016, 8(1): e2016007, DOI 10.4084/MJHID.2016.007
This article is available on PDF format at:


| This is an Open Access article distributed
under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by-nc/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |
|
Abstract Background and Objectives: Hepatitis
C virus (HCV) is a major health problem in Egypt with its prevalence
estimated to be 14.7% among the general population in 2008. Patients
receiving frequent blood transfusions like those with sickle cell
disease (SCD) are more exposed to the risk of acquiring HCV. IL28B gene
polymorphisms have been associated with spontaneous HCV clearance. This
study aims to determine the prevalence of HCV infection among children
with SCD and to investigate the relation between IL28B gene
polymorphisms and spontaneous HCV clearance. Methods: Seventy SCD patients were screened for HCV antibody. HCV-positive patients were tested for the level of HCV RNA using quantitative real-time PCR. IL28B polymorphisms (rs 12979860 SNP and rs 12980275 SNP) were detected using TaqMan QRT-PCR and sequence-specific primers PCR respectively. Results: Sixteen patients (23%) were HCV antibody positive, 9 of them (56.3%) had undetectable HCV RNA in serum, and 7 (43.7%) had persistent viremia. Genotypes CC/CT/TT of rs12979860 were found in 30 (42.9%), 29 (41.4%) and 11 (15.7%) patients and rs12980275 AA/AG/GG were found in 8 (11.4%), 59 (84.3%) and 3 (4.3%) patients. There was no significant difference in the frequency of IL28B (rs 12979860 and rs12980275) genotypes among HCV patients who cleared the virus and those with persistent viremia (p=0.308 and 0.724 respectively). Conclusion: Egyptian SCD patients have a high prevalence of HCV. Multi-transfused patients still exposed to the risk of transmission of HCV. IL28B gene polymorphismsare not associated with spontaneous clearance of HCV in this cohort of Egyptian children with SCD. |
Introduction
Egypt has the highest hepatitis C virus (HCV) prevalence worldwide.[1] The prevalence of HCV in Egypt is found to be 14.7% among the general population in the year 2008.[2]
HCV prevalence is even higher among hospitalized patients and special
clinical populations who have an increased risk of exposure to HCV like
multi-transfused patients, thalassemic patients, and patients on
hemodialysis.[3] The prevalence of HCV among Brazilian sickle cell disease (SCD) patients was found to be 14%.[4]
Again, a study from the USA found that 22% of SCD patients were
HCV-antibody positive. HCV positivity was most prevalent (58%) among
patients whom blood were drawn before 1992 when testing of all blood
donors in the USA became mandatory.[5] To date, the prevalence of HCV among SCD patients in Egypt is not known.
During the natural course of HCV, about 15% of patients show spontaneous viral clearance without treatment.[6]
HCV spontaneous clearance was defined as the lack of HCV-RNA detection
in the serum of the patient in the presence of a positive antibody
response and absence of antiviral therapy.[7] Factors
affecting viral clearance include age, gender, race, level of viremia,
alcohol intake, and HCV genotype. Genetic studies showed that single
nucleotide polymorphisms (SNPs) in the IL28B gene, which encodes
interferon (IFN)-λ-3 are associated with spontaneous HCV clearance.[8] Among the most significant SNPs were rs12979860 and rs12980275.[9-11]
The aim of this study is to determine the prevalence of HCV infection
and IL28B gene polymorphisms among Egyptian children with SCD and to
explore the possible relation between IL28B SNPs (rs12979860 and
rs12980275) and spontaneous viral clearance.
Patients and Methods
Patients and Methods.
Patients: This cross-sectional study included 70 Egyptian children with
SCD. The mean age of the patients was 10.2±4.5 years. They were 37
(52.8%) females and 33 (47.2%) males. Patients were consecutively
invited to participate in the study during their regular follow-up
visits at Pediatric Hematology Clinic, New Children Hospital, Cairo
University. The study was approved by the Ethical Committee of Kasr
Al-Ainy School of Medicine, Cairo University and patients were
recruited after informed consents were freely obtained from their
guardians.
In our resource-limited setting, SCD patients are not
routinely or regularly screened for HCV. Therefore, during recruitment
of the patients, their HCV status was not known, and none of the
patients received antiviral treatment. Patients’ records were reviewed
for the frequency of blood transfusion per year in the 12 months
preceding the enrollment. Laboratory testing included complete blood
count (CBC), reticulocyte count, Aspartate aminotransferase (AST) and
Alanine aminotransferase (ALT) levels. Table 1
shows clinical and laboratory data of the patients. Patients were
screened for HCV antibodies, and then quantification of HCV RNA in
serum for patients who were HCV antibody positive was done. To estimate
the frequencies of the IL28B genotypes in Egyptians, the SNPs
rs12979860 C/T SNP and rs12980275 A/G SNP were genotyped in the whole
group of SCD patients.
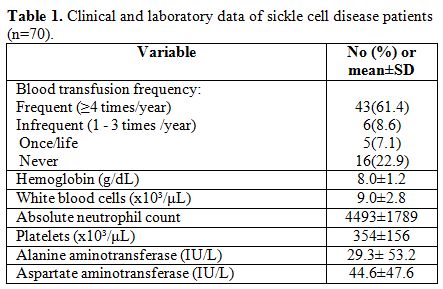 |
Table 1. Clinical and laboratory data of sickle cell disease patients (n=70). |
Methods:
Serum samples were tested for HCV antibody using a third generation
enzyme immunoassay (EIA) (Ortho HCV 3.0 ELISA test system, Ortho
Clinical Diagnostics Inc., Raritan, NJ, USA).
HCV RNA was
quantified using a commercial real-time RT-PCR assay (RealTime™ HCV,
Abbott Molecular Inc., Des Plaines, IL, USA) as specified by the
manufacturer. The detection limit was 12 IU/mL.
For detection of
IL28B polymorphisms, DNA was extracted using AxyPrep Blood Genomic DNA
Miniprep Kit (Axygen Biosciences, USA). IL28B polymorphisms rs 12979860
C/T genotyping was performed using the ABI TaqMan allelic
discrimination kit (Applied Biosystems, Foster City, California, USA)
as described before.[12]
The sequence of the used primers was: Forward 5'-TGCCTGTCGTGTACTGAACCA-3' and Reverse 5'-GAGCGCGGAGTGCAATTC-3'.
The
sequences of the Taqman probes were: Probe for the C allele
TGGTTCGCGCCTTC (VIC ™-labeled) and Probe for the T allele
CTGGTTCACGCCTTC (FAM ™-labeled).
The PCR reaction was carried out
in a total volume of 25µL. The following amplification protocol was
used: pre-incubation at 50°C for 2 minutes and then 95°C for 10
minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds,
and annealing/extension at 60°C for 1 minute.
The sequence specific primer -PCR was applied for genotyping IL28B SNP rs12980275A/G polymorphism as described previously.[13] A common reverse primer and two sequence-specific forward primers were used.
Gen
(antisense) 5'-ATGATCATAGCTCATTGCAGC-3' A allele-specific (sense)
5'-AGAAGTCAAATTCCTAGAAAC A-3' G allele-specific (sense)
5'-AGAAGTCAAATTCCTAGAAAC G-3'. Cycling condition was denaturation for 1
minute at 95°C, followed by 46 cycles of amplification; denaturation
for15 seconds at 95°C, annealing for 30 seconds at 50°C and extension
for 30 seconds at 72°C. In the last cycle, the extension was prolonged
to 7 minutes at 72°C. An amplification product of 393 bp was detected.
Statistical Methods:
Data were analyzed using Statistical Package for Social Sciences (SPSS)
version 21. Numerical data were expressed as mean ±standard deviation
(SD) and compared by student’s t-test. Qualitative data were expressed
as frequency and percentage and compared by Chi-square test or Fisher’s
exact test as appropriate. Odds ratio (OR) with its 95% confidence
interval (CI) were used for risk estimation. All p-values are
two-sided. P-values < 0.05 were considered significant.
Results
Screening of SCD patients for HCV antibody revealed that 16 (23%)
patients were positive for HCV antibodies. HCV antibody-positive
patients had significantly higher AST and ALT levels (p= 0.002 and
˂0.001, respectively) compared to HCV antibody-negative patients.
HCV
antibody-positive patients had a history of significantly more frequent
blood transfusion compared to HCV antibody-negative patients (p=
0.003). Patients who were receiving a frequent blood transfusion (≥4
times/year) had about 14-fold increased risk to be infected with HCV (Table 2).
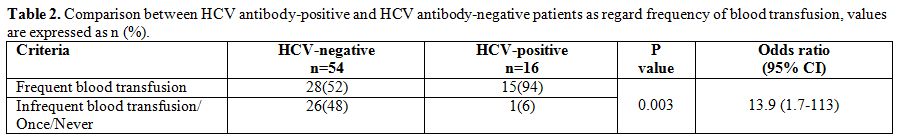 |
Table 2. Comparison between HCV antibody-positive and HCV antibody-negative patients as regard frequency of blood transfusion, values are expressed as n (%). |
Among HCV antibody-positive patients (n=16), 9 patients
(56.3%) had undetectable HCV RNA in serum (spontaneously cleared) and 7
patients (43.7%) were non-cleared. HCV non-cleared patients had
significantly higher AST and ALT levels (p= 0.012 and 0.023,
respectively) compared to patients with spontaneous virus clearance.
Genotyping
of SCD patients for IL28B rs 12979860 C/T polymorphism revealed that;
the wild type (CC) was detected in 30 patients (42.9%), heterozygous
genotype (CT) was detected in 29 patients (41.4%), and the homozygous
genotype (TT) was detected in 11 patients (15.7%). The frequency of the
wild allele (C) was 0.64, and the frequency of the mutant allele (T)
was 0.36.
Genotyping of SCD patients for IL28B rs12980275 A/G
polymorphism revealed that; the wild type (AA) was detected in 8
patients (11.4%), heterozygous genotype (AG) was detected in 59
patients (84.3%), and the homozygous genotype (GG) was detected in 3
patients (4.3%). The frequency of the wild allele (A) was 0.54, and the
frequency of the mutant allele (G) was 0.46. To analyze the impact of
polymorphisms of IL28B gene on HCV clearance, the prevalence of
rs12979860 C/T and rs12980275 A/G polymorphisms was compared between
patients spontaneously clearing and patients not clearing HCV (Table 3), and no statistically significant difference was found between the two groups as regard prevalence of polymorphisms.
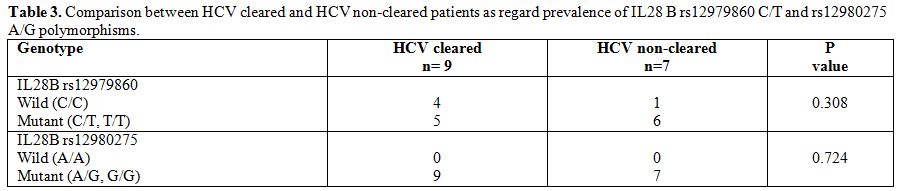 |
Table 3. Comparison between HCV cleared and HCV non-cleared patients as regard prevalence of IL28 B rs12979860 C/T and rs12980275 A/G polymorphisms. |
Discussion
The prevalence of HCV among SCD patients in Egypt is not known. The
present study is the first to investigate HCV prevalence among Egyptian
SCD patients that is found to be 23%. Previous studies in Egypt
revealed that HCV prevalence is high among all special clinical
population groups like hemodialysis patients (35%),[14] hemophilic children (40%),[15] multi-transfused thalassemic patients (40.5%),[16] non-Hodgkin's lymphoma patients (43%)[17] and hospitalized patients referred for bone marrow examination (42%).[18]
Hospitalization, repeated blood transfusions, invasive procedures,
injections and shared dialysis machines are common risk factors for HCV
infection among these group of patients. In this study, HCV
antibody-positive patients received blood transfusion more frequently
than HCV antibody-negative patient. Although the incidence of
transfusion-acquired infections has significantly decreased in recent
years because of more effective donor screening methods, the risk is
still present, especially in multi-transfused patients.[19]
At Cairo University blood bank, donated blood is routinely screened for
HCV using a third generation EIA. The more sensitive molecular methods
utilizing nucleic acid amplification technology are not used due to
limited resources. Previous studies have shown that the number of
transfusions was directly correlated with the HCV antibody positivity.
Patients who received 10 or more blood units had significantly higher
incidence of anti-HCV markers than those receiving less than ten units
of blood products.[5] In the current study, patients
who were receiving a frequent blood transfusion (≥4 times/year) had
about 14-fold increased risk to be infected with HCV.
Hepatitis
C leads to persistent infection in a high proportion of infected
individuals, and can progress to chronic liver disease, cirrhosis, and
hepatocellular carcinoma. HCV spontaneous clearance was defined as the
lack of HCV-RNA detection in the serum of the patient in the presence
of a positive antibody response and absence of antiviral therapy.[7]
In the current study, 56.3% of HCV positive patients spontaneously
cleared the virus. During the natural course of HCV, about 15% of
patients showed spontaneous viral clearance without treatment.[6]
As observed in the current study, higher rates of spontaneous
resolution have been found in children. In a prospective study
including 67 patients with chronic HCV, infection due to blood
transfusion at a mean age of 2.8 years, the infection resolved in 30
patients (45%) after a mean follow-up of 20 years; of the remaining 37
patients, only 1 had abnormal liver enzymes, and only 3 showed signs of
histologic damage.[20] Factors responsible for
age-related differences in the clinical course of chronic HCV infection
may include, structural and/or immunologic differences between children
and adults including less availability of antioxidizing agents and fat
infiltration of adult liver.[21]
Ethnic differences in the frequency of virus clearance suggest that host genetic variation may have an impact on HCV clearance.[22]
Genetic studies showed that genetic variation in the IL28B gene, which
encodes IFN-λ-3 is associated with spontaneous HCV clearance.[8]
Other studies have reported associations of SNPs in IL28B gene with
response to antiviral therapy. Among the most significant SNPs were
rs12979860 and rs12980275.[9-11] In the current study,
the allele frequency at the rs12979860 SNP was 64% for the wild allele
C and 36% for the mutant allele T. The frequency of the wild allele in
this study is similar to that detected in a previous Egyptian study
(67%),[23] in another North African (Moroccan) population (68%)[12] and in some European populations (60-70%),[8] but higher than that found in southern African populations (23-40%) and less than in Asian population (75-98%).[8]
The allele frequency at the rs12980275 SNP was 54% for the wild allele
A and 46% for the mutant allele G. This wild allele frequency is
similar to that among Caucasian population (52%)[13] but less than that detected in previous studies including Saudi population (62%)[24] and European population (62.6%).[25]
Our
results show that IL28B gene polymorphisms are not associated with
spontaneous resolution of HCV in this group of children with SCD.
However, the generalization of these preliminary observations is
limited by the small sample size. The association between SNP
(rs12979860) of the IL28B gene and the outcome of HCV infection was
first described in 2009. Investigators found that the CC genotype of
the rs12979860 SNP was associated with an improved response to
treatment of adult patients with HCV independent of their ethnicities.[9]
Since that, multiple studies have confirmed and reinforced the
association between the C allele of the rs12979860 SNP and both
spontaneous and treatment-induced clearance of HCV in adults.[26]
Fewer
data are available regarding the effect of rs12980275 SNP of IL28B gene
on HCV outcome. A genome-wide association study in 2009 found that the
A allele of the rs12980275 SNP was associated with good response to
treatment in adult Japanese patients with HCV.[11] A subsequent study confirmed the association between rs12980275 SNP and higher response to treatment in adults with HCV.[24]
In
children, data about the relation between polymorphisms in IL28B gene
(rs12979860 and rs12980275) and spontaneous clearance of HCV is
limited. In contrast to our results and in agreement with that already
demonstrated in adults, two studies published in 2011 demonstrated for
the first time that the C allele of rs12979860 SNP of the IL28B is
associated with spontaneous clearan ce of HCV in children.[27,28] These preliminary results have been confirmed in a subsequent study including a larger cohort of Italian children with HCV.[29]
To
our knowledge, no previous study addressed the relation between
rs12980275 SNP of IL28B gene and spontaneous resolution of HCV in
children.
The mechanism behind the association of genetic
variations in the IL28B gene and spontaneous clearance of HCV may be
related to the host innate immune response. IL28B encodes IFN-λ-3,
which is involved in viral control, including HCV.[30]
Both IFN-α and IFN-λ-3 bind to cell-surface receptors leading to
induction of interferon stimulating genes, a mechanism by which IFNs
suppress viral infections.[30-32]
Conclusions
The present study revealed that the prevalence of HCV infection among Egyptian patients with SCD is considerably high. Despite more-effective donor screening methods, blood transfusion still carries a risk of transmission of HCV, especially in multi-transfused patients. Efforts should be made to implement a more sensitive molecular method for screening of blood products for HCV. In the current study, more than half of HCV positive patients show spontaneous viral clearance. Polymorphisms in the IL28B may not be associated with spontaneous clearance of HCV infection among this group of children with SCD. This study is limited by the small number of HCV positive patients. Before generalization of our results, larger studies are required to assess better the impact of genetic variation in IL28B gene on HCV outcome in children.
References































[TOP]