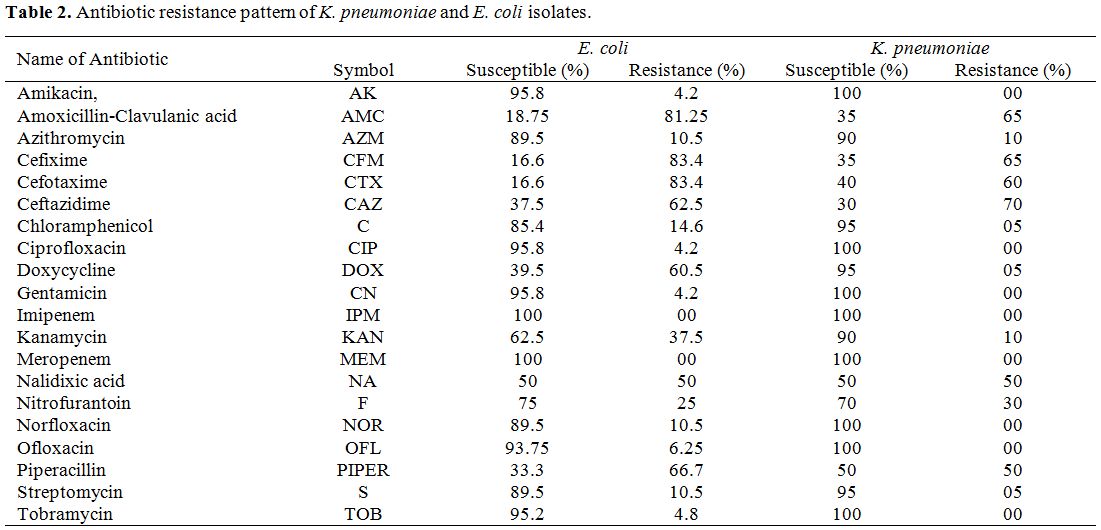
Antimicrobial susceptibility testing. According to the guidelines of the Clinical and Laboratory Standards Institute (CLSI), the isolates were screened by the disc diffusion method (Kirby-Bauer disc diffusion method) on Mueller-Hinton agar (MHA) plates in order to test their antimicrobial susceptibility.[11] The utilized antimicrobials included Amoxicillin+Clavulanic acid (20+10μg), Amikacin (10 μg), Azithromycin (15μg), Cefixime (5μg), Cefotaxime (30μg), Chloramphenicol (30μg), Ceftazidime (30μg), Ciprofloxacin (10μg), Doxycycline (30μg), Imipenem (10μg), Gentamicin (10μg), Kanamycin (30μg), Nalidixic acid (30μg), Meropenem (10μg), Nitrofurantoin (100μg), Norfloxacin(10μg), Ofloxacin (5μg), Streptomycin (25μg), Piperacillin (100μg), and Tobramycin (10μg).
Testing for production of ESBL (MDDST). Using a disc of Amoxicillin-Clavulanate (20/10 μg) with four cephalosporins of Ceftriaxone, 3GC-Cefotaxime, 4GC-Cefepime, and Cefpodoxime, the Modified Double Disc Synergy Test (MDDST) was employed to test all strains in terms of their production of Extended Spectrum Beta-Lactamase (ESBL). A lawn culture belonging to the organisms was created on a Mueller-Hinton agar plate following the recommendations by CLSI.[11] A disc that contained Amoxicillin-Clavulanate (20/10 μg) was put in the middle of the plate. The 3GC and 4GC discs were placed respectively 15mm and 20mm center-to-center apart from the center of the amoxicillin-clavulanate disc.[12] Any increase or distortion in the zone toward the Amoxicillin-Clavulanate disc was regarded positive for the production of ESBL. According to CLSI guidelines, the combined disc test was used to confirm ESBL production.
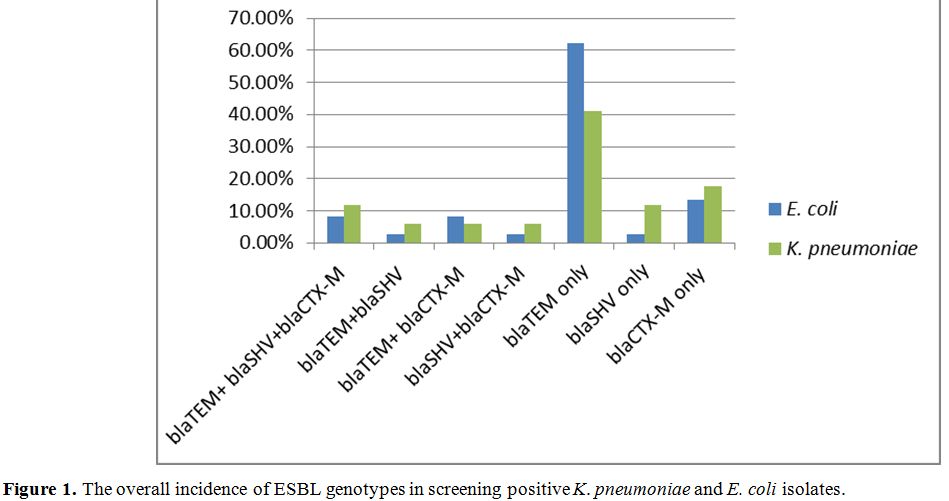
Detection of ESBL genotypes by multiplex PCR amplification. Using the method utilized by Monstein et al. (2007) with slight modifications, multiplex PCR was employed to examine the positive isolates in the initial screening test for ESBL production for the existence of blaSHV, blaCTX-M, and blaTEM genes.[13] Freshly cultured isolates bacteria were used to prepare template deoxyribonucleic acid (DNA) was prepared using PrestoTM Mini gDNA bacterial kit. All reactions of PCR were conducted by utilizing 2 μl DNA template (density of 10 ng/µl), the Master Mix consisting of 3 mM MgCl2, 0.2% Tween® 20, 20 mM Tris-HCl pH 8.5, (NH4)2S04, 0.4 mM of each dNTP, 0.4 μM of each primer, and 0.2 units/µl Ampliqon Taq DNA polymerase. The conditions of polymerase chain reaction amplification were set up as follow: primary denaturation step for 10 minutes at 95°C; 30 denaturation cycles for 30 seconds at 94°C, annealing 30 seconds at 60°C for, extension for 2 minutes at 72°C, and a final extension step for 10 minutes at 72°C. Using agarose gel electrophoresis, size separation PCR amplicons were utilized to detect respective genes (Table1).
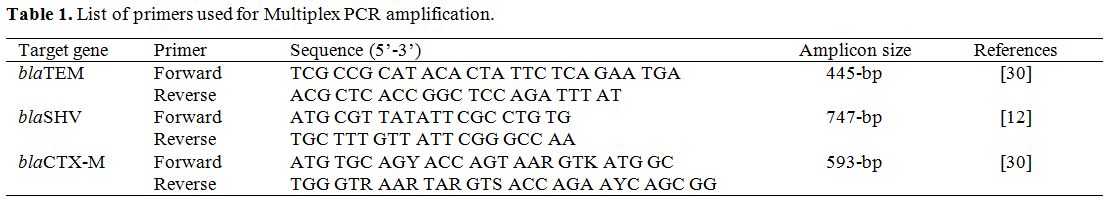 |
Table 1. List of primers used for Multiplex PCR amplification. |