Kursat Ozdilli1, Mustafa Pehlivan2,3, Istemi Serin2, Fatma Oguz Savran4, Ayse Gaye Tomatir5 and Sacide Pehlivan4.
1 Medipol University, Faculty of Medicine, Department of Medical Biology.
2 University of Health Sciences, Istanbul Training and Research Hospital, Hematology Clinic.
3 Gaziantep University, Faculty of Medicine, Department of Hematology.
4 Istanbul University, Istanbul Faculty of Medicine, Department of Medical Biology.
5 Pamukkale University, Faculty of Medicine, Department of Medical Biology.
Correspondence to: Istemi Serin, MD. University of Health
Sciences, Istanbul Training and Research Hospital, Department of
Hematology, Org.Nafiz GURMAN Cad, 34098, Fatih, Istanbul. Tel: 0090 532
3172393. E-mail:
serinistemi@hotmail.com
Published: March 1, 2021
Received: November 14, 2020
Accepted: February 12, 2021
Mediterr J Hematol Infect Dis 2021, 13(1): e2021020 DOI
10.4084/MJHID.2021.020
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
To the editor
Chronic
myeloid leukemia (CML), which is characterized by the overproduction of
mature cells in the granulocytic series, is included in the group of
chronic myeloproliferative neoplasms.[1] It is the
first disease ascertained as due to a specific chromosomal anomaly
emerging from a reciprocal translocation between chromosomes 9 and 22.
A chimeric gene denominated as the Philadelphia (Ph) chromosome is the
product of the fusion of the Abelson oncogene (ABL) from chromosome
9q34 with the breakpoint cluster region (BCR) on chromosome 22q11.2, t
(9;22)(q34;q11.2).[1]
New approaches are tried to
be developed in evaluating the prognosis and treatment response. DNA
repair mechanisms create a new study area for CML and constitute the
subject of our study. There are more than 100 known DNA repair genes.
Polymorphisms and/or functional gene variants occurring in these genes
with environmental factors increase the cancer tendency by disrupting
the DNA repair mechanism.[2]
The ERCC2 (excision
repair cross complementation group 2) gene acts on nucleotide excision
repair (NER) and is located in the 13.3rd district of the Q part in the 19th chromosome.[5]
Polymorphisms in the ERCC2 gene provide information about DNA repair
capacity and cancer risk. ERCC2 repair gene polymorphisms are
significantly associated with breast, colorectal, pancreatic, bladder,
lung, esophageal cancers and hematological malignancies.[3-5]
The
XRCC1 (X-ray repair cross-complementing group 1) gene is one of the BER
genes and is located in the 13.2 district of the q part in the 19th chromosome. This gene has 17 exons required to synthesize DNA proteins, including DNA polymerase.[5]
Polymorphisms in the XRCC1 repair gene have been investigated,
especially in colorectal, breast, pancreatic, head and neck, lung,
prostate, and skin cancers.
The DNA repair protein XRCC4, also
known as X-ray repair cross-complementing protein 4, is a protein
encoded in humans by the XRCC4 gene. XRCC4, an important non-homologous
splice repair gene, acts as an essential scaffold protein between this
complex and DNA Ligase IV in the DNA double-stranded break repair
pathway process.[6]
In our study, we aimed to
examine the effect of ERCC2 (751), XRCC1 (399), XRCC4-Intron 3, and
XRCC4 (-1394) gene polymorphism on CML, prognosis, and treatment
response in patients.
Patients and Methods
Sixty-two
(62) CML patients, diagnosed and followed up in the Gaziantep
University Hematology Clinic between January 2008 - January 2016, and a
control group of 70 healthy people were included in the study. In
addition to demographic data such as age and gender, initial Sokal risk
scores, presence of splenomegaly, initial laboratory values
(hemoglobin, leukocytes, platelets), treatment preferences (imatinib or
interferon alfa), responses at 18 months according to European Leukemia
Net (ELN) criteria, mortality, presence of any events, chromosome
abnormalities, overall survival (OS) and event-free survival (EFS)
durations (months) were recorded. The median age of all 62 patients
included in the study was 41 (range: 20-74)
DNA isolation from
peripheral blood leukocytes of CML patients and controls was performed
using the saline precipitation method (Miller et al.).[7]
ERCC2, XRCC1, XRCC4-Intron 3, XRCC4 (-1394) gene polymorphism genotypes
were analyzed by Polymerase chain reaction (PCR) and/or Polymerase
chain reaction-restriction fragment length polymorphism (PCR-RLFP)
method.
SPSS for Windows (version 13.0; SPSS, Chicago, IL)
software was used for data analysis. Logistic regression analysis was
used to determine the statistical significance of the differences
between control groups and patients. The odds ratios (OR) and 95%
confidence intervals were used for this analysis. The X2
test was used to compare the differences between the patient groups and
the control group's DNA Repair Gene XRCC4 variable number tandem repeat
(VNTR) at intron 3 and -1394), XRCC1, ERCC2 allele frequency. Fisher's
test was used as needed. P values <0.05 were considered to indicate
statistical significance. The Kaplan-Meier method was used to estimate
the survival probabilities and the log-rank test to compare
differences. The significance of risk factors was confirmed by applying
The Cox stepwise regression analysis. In the multivariate analysis, the
stepwise (backward) eliminated variables were used with a significance
of less than 10%.
Results
Looking
at the molecular responses of the patients at 18 months, 42 were in the
optimal (67.7%), 13 were in the warning (21%), and 7 (11.3%) were in
the failure group. End-of-study mortality was 3.2% with two patients (Table 1).
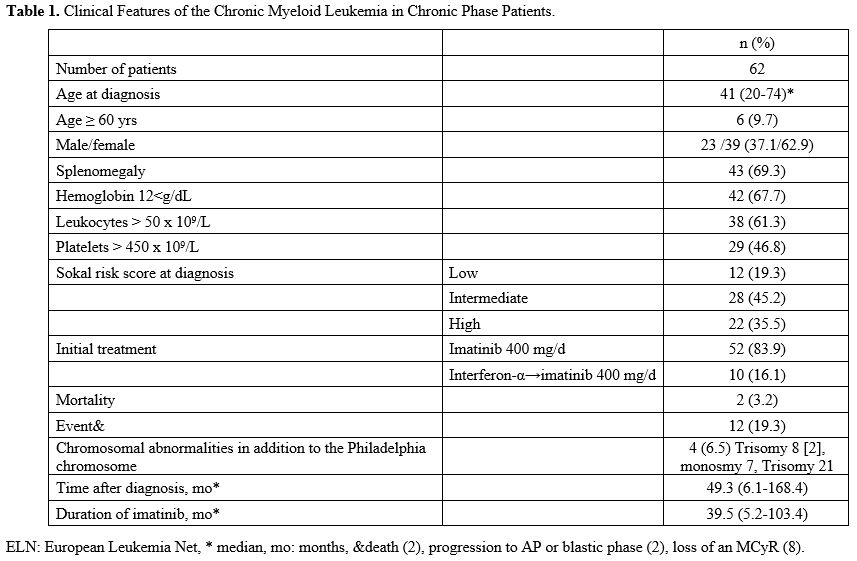 |
Table
1. Clinical Features of the Chronic Myeloid Leukemia in Chronic Phase Patients.
|
Twelve
(12) of the patients experienced any "event" (19.3%). Two of them were
exitus (3.2%), 2 of them showed a progression to accelerated phase or
blastic phase (3.2%), and 8 of them lost major molecular response (MMR)
(12.9%). The median follow-up period was 49.3 months (6.1-168.4), and
the median use of imatinib was 39.5 months (5.2-103.4) (Table 1).
When
the genotype differences for ERCC2, XRCC1, and XRCC4 (-1394) between
CML and healthy controls were analyzed, there was no statistically
significant difference found between the two groups (p> 0.05). When
XRCC4-Intron 3 was examined, it was observed that there was a
significant statistical difference in DD and II genotypes between CML
and the control group (p = 0.018, p = 0.028). It was also observed that
the DD genotype was 7.299 times protective factor for CML, and patients
with II genotype have 2.379 times increased risk of CML (Table 2).
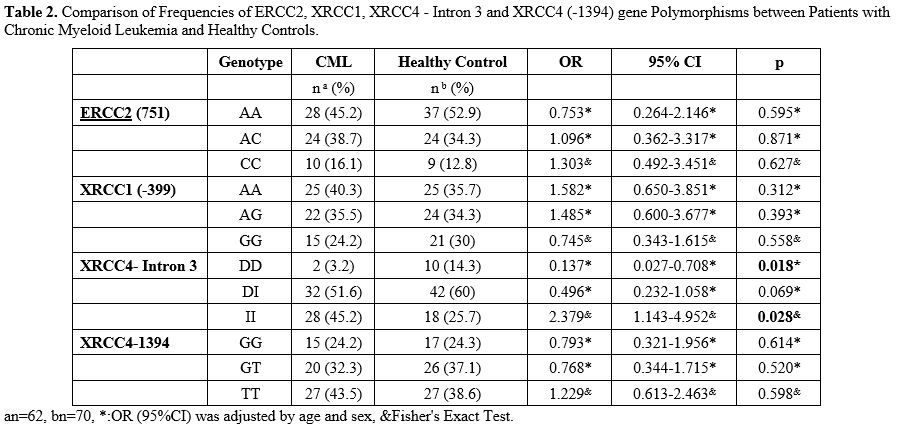 |
Table
2. Comparison of Frequencies of ERCC2, XRCC1, XRCC4 - Intron 3 and
XRCC4 (-1394) gene Polymorphisms between Patients with Chronic Myeloid
Leukemia and Healthy Controls.
|
Four
different factors were found to be statistically significant for EFS.
Young age (<60) (p = 0.020), absence of splenomegaly (p = 0.011),
presence of low Sokal risk score at initial diagnosis (p = 0.0148) and
presence of XRCC1 GG genotype (p = 0.033) were statistically
significant for better EFS (Table 3, Figure 1).
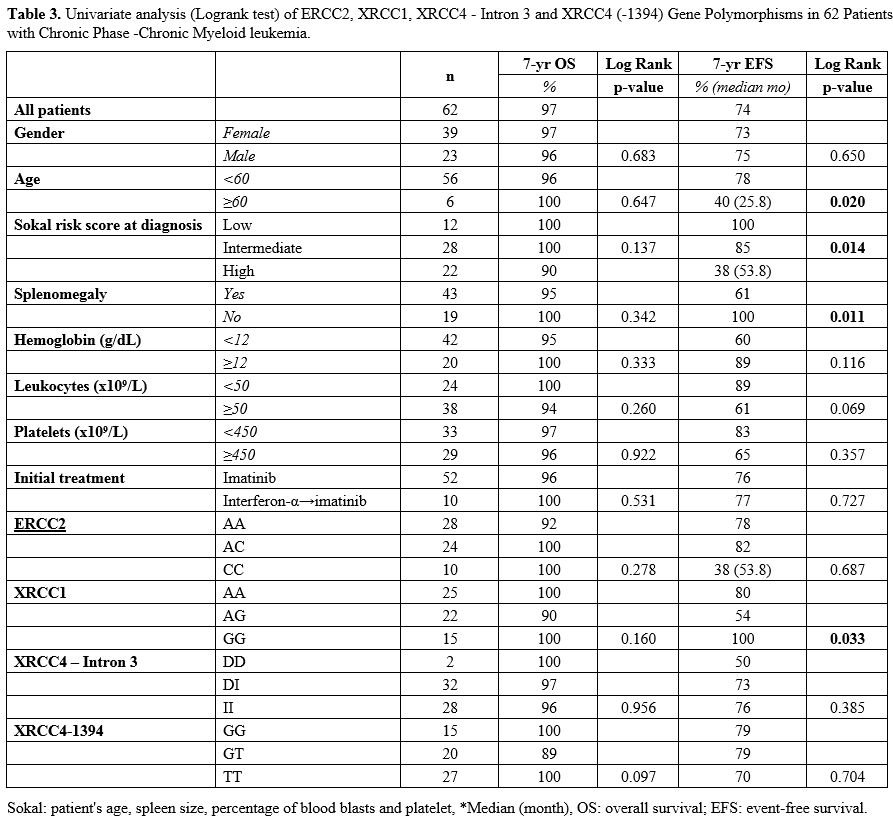 |
Table 3. Univariate
analysis (Logrank test) of ERCC2, XRCC1, XRCC4 - Intron 3 and XRCC4
(-1394) Gene Polymorphisms in 62 Patients with Chronic Phase -Chronic
Myeloid leukemia. |
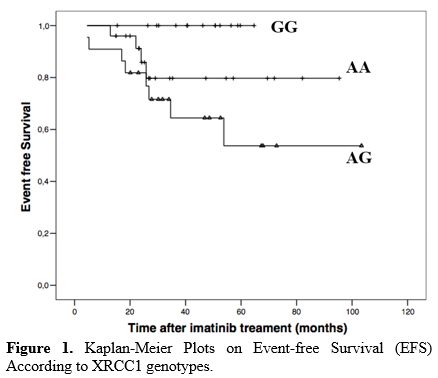 |
Figure
1. Kaplan-Meier Plots on Event-free Survival (EFS) According to XRCC1 genotypes. |
Discussion
The
literature data about DNA repair mechanisms in hematological
malignancies is limited. In a study conducted by Salimizand et al.,[8]
simultaneous effects of polymorphism of three separate DNA repair genes
were investigated on CML development. T allele of ABCB1 C3435T, T
allele of XRCC1 Arg194Trp, and C allele of ABCG2 C421A polymorphisms
were significantly higher CML patients compared to controls. TT
genotype of ABCB1 and XRCC1 has been associated with a higher risk of
developing CML.
In a meta-analysis, Wang et al.[9]
examined the relationship between the Arg399Gln single nucleotide
polymorphism (SNP) in the XRCC1 gene and the risk of leukemia. No
association was found between XRCC1 and CML. Among the articles
discussed in this meta-analysis, 2 of them were directly related to
CML: Deligezer et al.[10] investigated the
association of XRCC1 gene polymorphism Arg399Gln with CML and could not
obtain a significant difference among patient groups. Similarly, in our
study, no relationship was found between this polymorphism and CML.
Annamaneni et al.[11] studied the XRCC1 effect on CML
and polymorphisms of XRCC1, codon 399, 280 and 194; similarly, no
significant difference was detected.
Dhangar et al.[12]
investigated the correlation between clinical response to therapy
between CML and XRCC1 rs1799782, rs25487, and ERCC2 rs13181
polymorphisms; no significant relationship was found. Banescu et al.[13]
also examined the relationship between CML and XRCC1 Arg399Gln,
Arg280His, Arg194Trp, XRCC3 r241Met, and ERCC2 Lys751Gln polymorphisms
and showed that the ERCC2 Lys751Gln genotype increases the risk of CML.
Ozcan et al.[14] investigated the place of
ERCC2 and XRCC1 gene polymorphisms in different hematological
malignancies. In his study, he showed that a decrease in the Gln / Gln
genotype and the Gln allele in the ERCC2 codon 751 and XRCC1 codon 399
polymorphisms play a protective role in AML, and an increase in Lys/Lys
genotype in acute leukemia was associated with early relapse.
Joshi et al.[15]
studied XRCC1 and ERCC2 polymorphisms in myelodysplastic syndrome
(MDS), showing that the progression of MDS to AML be the result of the
gradual accumulation of DNA mutations that create a defect in DNA
repair. DNA repair gene XRCC1 (Arg280His) (p = 0.05) and ERCC2
(Lys751Gln) (p = 0.01). Polymorphisms were significantly higher in MDS
patients compared to controls. There was a significant difference
between RAEB I and XRCC1, being XRCC1 polymorphisms strongly associated
with the advanced MDS subgroup.
In our study, different from the
other two main studies, we also had the opportunity to evaluate XRCC4
and CML's relationship. When the genotype differences between CML and
healthy control groups were statistically analyzed, no statistically
significant difference could be found between them. However, when
XRCC4-Intron 3 was examined, it was seen that there was a significant
statistical difference in DD and II genotypes between CML and the
control group. Additionally, it was observed that the DD genotype was
7.299 times protective factor for CML, and patients with II genotype
have 2.379 times increased risk of CML.
The study also had some
limitations. The most important limitation is the small patient
population. It is thought that significant results can be obtained in
terms of disease parameters and prognosis with the data in a larger
patient group. Besides, only imatinib and interferon-related treatment
results could be evaluated in our study. It would be more meaningful in
terms of the literature to conduct a study on 2nd generation tyrosine kinase inhibitors (TKIs) throughout a broader period.
References
- Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M.
Philadelphia chromosome-positive leukemias: from basic mechanisms to
molecular therapeutics. Ann Intern Med. 2003 May 20;138(10):819-30. https://doi.org/10.7326/0003-4819-138-10-200305200-00010 PMid:12755554
- Das-Gupta
EP, Seedhouse CH, Russell NH. DNA repair mechanisms and acute
myeloblastic leu-kemia. Hematol Oncol. 2000 Sep;18(3):99-110. https://doi.org/10.1002/1099-1069(200009)18:3<99::AID-HON662>3.0.CO;2-Z
- Taylor
EM, Broughton BC, Botta E et al. Xeroderma pigmentosum and
trichothiodystrophy are as-sociated with different mutations in the XPD
(ERCC2) repair/transcription gene. Proc Natl Acad Sci U S A. 1997 Aug
5;94(16):8658-63. https://doi.org/10.1073/pnas.94.16.8658 PMid:9238033 PMCid:PMC23065
- Artac
M, Bozcuk H, Pehlivan S et al (2010). The value of XPD and XRCC1
genotype polymor-phisms to predict clinical outcome in metastatic
colorectal carcinoma patients with irinotecan-based regimens. J. Cancer
Res. Clin. Oncol. 136: 803-809. https://doi.org/10.1007/s00432-009-0720-3 PMid:19908066
- Chacko
P, Rajan B, Joseph T, Mathew BS, Pillai MR. Polymorphisms in DNA repair
gene XRCC1 and increased genetic susceptibility to breast cancer.
Breast Cancer Res Treat. 2005;89(1):15-21. https://doi.org/10.1007/s10549-004-1004-x PMid:15666192
- Lu
J, Wang XZ, Zhang TQ et al. Prognostic significance of XRCC4 expression
in hepatocellular carcinoma. Oncotarget. 2017 Sep 28;8(50):87955-87970.
https://doi.org/10.18632/oncotarget.21360 PMid:29152133 PMCid:PMC5675685
- Miller
SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting
DNA from human nucleated cells. Nucleic Acids Res. 1988 Feb
11;16(3):1215. https://doi.org/10.1093/nar/16.3.1215 PMid:3344216 PMCid:PMC334765
- Salimizand
H, Amini S, Abdi M, Ghaderi B, Azadi NA. Concurrent effects of ABCB1
C3435T, ABCG2 C421A, and XRCC1 Arg194Trp genetic polymorphisms with
risk of cancer, clinical out-put, and response to treatment with
imatinib mesylate in patients with chronic myeloid leukemia. Tumour
Biol. 2016 Jan;37(1):791-8. https://doi.org/10.1007/s13277-015-3874-4 PMid:26250462
- Wang
F, Zhao Q, He HR et al. The association between XRCC1 Arg399Gln
polymorphism and risk of leukemia in different populations: a
meta-analysis of case-control studies. Onco Targets Ther. 2015 Nov
6;8:3277-87. https://doi.org/10.2147/OTT.S92752 PMid:26609240 PMCid:PMC4644162
- Deligezer
U, Akisik EE, Dalay N. Lack of association of XRCC1 codon 399Gln
polymorphism with chronic myelogenous leukemia. Anticancer Res. 2007
Jul-Aug;27(4B):2453-6.
- Annamaneni S,
Gorre M, Kagita S et al. Association of XRCC1 gene polymorphisms with
chronic myeloid leukemia in the population of Andhra Pradesh, India.
Hematology. 2013 May;18(3):163-8. https://doi.org/10.1179/1607845412Y.0000000040 PMid:23320983
- Dhangar
S, Shanbhag V, Shanmukhaiah C et al. Lack of association between
functional polymor-phism of DNA repair genes (XRCC1, XPD) and clinical
response in Indian chronic myeloid leuke-mia patients. Mol Biol Rep.
2019 Oct;46(5):4997-5003. https://doi.org/10.1007/s11033-019-04950-0 PMid:31286393
- Bănescu
C, Trifa AP, Demian S, Benedek Lazar E, Dima D, Duicu C, Dobreanu M.
Polymorphism of XRCC1, XRCC3, and XPD genes and risk of chronic myeloid
leukemia. Biomed Res Int. 2014;2014:213790. https://doi.org/10.1155/2014/213790 PMid:24955348 PMCid:PMC4052066
- Özcan,
A, Pehlivan, M, Tomatir, A et al. (2011). Polymorphisms of the DNA
repair gene XPD (751) and XRCC1 (399) correlates with risk of
hematological malignancies in Turkish population. African Journal of
Biotechnology. 10. 8860-8870. 10.5897/AJB10.1839. https://doi.org/10.5897/AJB10.1839
- Joshi
D, Korgaonkar S, Shanmukhaiah C, Vundinti BR. Association of XPD
(Lys751Gln) and XRCC1 (Arg280His) gene polymorphisms in myelodysplastic
syndrome. Ann Hematol. 2016 Jan;95(1):79-85. https://doi.org/10.1007/s00277-015-2528-3 PMid:26482462
[TOP]



