Muhamad R. Abdel Hammed1*, Sherein G. Elgendy2*, Mohamed A. El-Mokhtar2, Douaa Sayed3, Samar M. Mansour3 and Abeer M. Darwish3.
1 Internal
Medicine Department and Hematology Unit, Assiut University Hospitals
and South Egypt Cancer Institute Bone Marrow Transplantation Unit,
Assiut University, Assiut, Egypt.
2 Department of Medical Microbiology and Immunology, Faculty of Medicine, Assiut University, Assiut, Egypt.
3 Department of Clinical Pathology, South Egypt Cancer Institute, Assiut University, Assiut, Egypt.
* Both author equally contributed to the work.
Correspondence to:
Muhamad R. Abdel Hameed, MD. Department of Internal Medicine
& Hematology Unit, Assiut University Hospitals and Bone Marrow
Transplantation Unit, South Egypt Cancer Institute, Assiut
University, Assiut, Egypt. Tel: (+2) 01097510010, Fax:
+088-2080278. E-mail:
dr.muhamadramadan@yahoo.com Sherein
G. Elgendy, Ph.D. Department of Medical Microbiology and Immunology,
Faculty of Medicine, Assiut University, Egypt. Tel.: (+2) 01021887728,
Fax: +088-2080278. E-mail:
Shereinelgendy@yahoo.com
Published: March 1, 2022
Received: October 29, 2021
Accepted: February 8, 2022
Mediterr J Hematol Infect Dis 2022, 14(1): e2022022 DOI
10.4084/MJHID.2022.022
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background: Invasive
fungal infections (IFIs) are important cause of mortality in acute
myeloid leukemia (AML) patients on treatment with intensive induction
chemotherapy. Toll-like receptors, mainly Toll-like receptors 2 and 4
(TLR2 and TLR4), play a considerable role in the host defense against
microorganisms. The involvement of TLR signaling in modulation of
innate immune response is extensively discussed, but the TLR
expressions profiling on adaptive immune cells are not specified. Also,
the expressions of TLRs and their association with the occurrence of
IFIs in patients with AML are not studied. So, the novel aim of this
study was to investigate the associations between the T-lymphocyte
expression of TLR2 and TLR4 and the occurrence of IFIs in AML patients
treated with intensive induction chemotherapy.
Materials and Methods: One
hundred twenty two newly diagnosed AML patients were evaluated. The
laboratory diagnostic techniques for IFIs include culture, microscopic
examination, histopathology, galactomannan assay and PCR. The
expressions of TLR2 and TLR4 were analyzed by flow cytometry. The
Control group included 20 age and sex-matched individuals. Results: There was a significant increase in the expression of TLR4 in AML patients with IFI compared to healthy controls (p=0.001).
TLR2 and TLR4 expressions increased significantly in AML patients with
mixed fungal and bacterial infection compared to healthy controls (p= 0.002 and p=0.001, respectively).
Conclusion: TLRs
expressions could be important biological markers for the occurrence of
IFI in non-M3 AML patients after intensive induction chemotherapy.
|
Introduction
Acute
myeloid leukemia (AML) represents the hematologic malignancy with the
highest risk of invasive fungal infections (IFIs). The overall
mortality rate in AML patients due to fungal infections was improved in
recent years to 20%-30%.[1] IFIs represent a
considerable clinical problem due to the high costs of the prophylaxis
and treatment of fungal infections in limited resource localities.[2]
Multiple
risk factors can predispose AML patients to develop fungal
infections including old ages, pulmonary comorbidities, duration of
neutropenia, relapse/refractory disease, intense chemotherapy, and a
high dose of steroids.[3]
The Infectious Diseases
Working Party of the German Society of Hematology and Medical Oncology
(AGIHO) postulates that prolonged severe neutropenia in AML patients
(<500 cells/μL of at least 8 days) post- induction/consolidation
chemotherapy or allogeneic stem cell transplantation are considered as
individuals at high-risk for IFI.[4]
Diagnosis of
IFI is challenging, particularly in AML patients as symptoms can be
absent or subtle. Fever may be the only sign. Thrombocytopenia and
coagulopathy due to the underlying cause and chemotherapy may impair
the ability to tissue biopsy which is the preferred method for
diagnosis establishment.5 The European Organization for Research and
Treatment of Cancer/Invasive Fungal Infections Cooperative Group
(EORTC) and the National Institute of Allergy and Infectious Diseases
Mycoses Study Group (MSG) defining IFI as proven, probable, and
possible infections.[5,6]
Recognition of fungi
by immune cells is mediated through pattern recognition receptors
(PRRs); like Toll-like receptors (TLRs) and C-type lectins (CLRs).
Binding of fungal pathogen-associated molecular patterns (PAMPs) to
PRRs triggers phagocytes to the infection site, microbial killing, and
dendritic cells (DCs) activation.[7,8]
Toll-like receptors are widely expressed on myeloid cells of innate immune system, such as macrophages, DCs.[9]
TLR signaling in DCs triggers a maturation program that increases their
ability to prime naïve T cells through up-regulation of MHC and
co-stimulatory molecules and expression of pro-inflammatory cytokines,
such as TNF-α, IL-1, and IL-6.[10]
TLRs have
been considered traditionally to play an important role in the innate
immune system. However, other few studies have found that TLRs are also
expressed on various adaptive immune cells, such as B cells,[11] CD4+ and CD8+ T cells,[12] and the CD4+CD25+ regulatory T cell population.[13]
Two studies, sorted CD4+CD45high T cells from C57/BL6 (B6) mice were
found to express TLR1, 2, 3, 6, 7 and 8, but low levels of TLR 4, 5 and
9 mRNA.[14] Naïve CD8+ T cells from B6 mice were reported to express mRNA for TLR1, 2, 6 and 9 but not TLR4.[15] Naïve CD4+ T cells from BALB/c mice express mRNA for TLR3, 4, 5 and 9.[16]
Thus, the involvement of TLR signaling in modulation of immune response
is not limited to innate immune cells, but also modulate cellular and
humoral adaptive immunity. TLR2 and TLR4 are two of the most studied
TLRs to have an important role in the recognition of both bacterial and
fungal pathogens.17 So, we have focused on the associations between
T-lymphocyte expression of TLR2 and TLR4 and the occurrence of IFIs in
AML patients which remains unclear.
Materials and Methods
Ethics Statement.
This study was approved by the Regional Ethical Committee in South
Egypt Cancer Institute (SECI), Assiut University, in accordance with
the provisions of the Declaration of Helsinki. Informed written consent
obtained from all participants before enrolment.
Study Design and Setting.
This study was performed at Clinical Hematology Unit, Internal Medicine
Department, Assiut University Hospital, and South Egypt Cancer
Institute (SECI), Assiut University, Egypt. All newly diagnosed AML
patients (aged more than 18 years old), admitted in the duration from
October 2017 to July 2020 were enroll in this study. The diagnosis was
performed according to the WHO criteria for AML. [18] The intensive induction chemotherapy was (idarubicin 12 mg/m2 per day for 2–3 days, and cytarabine 100 mg/m2/day
for 5–7 days). Patients received prophylactic treatment during the
period of neutropenia following chemotherapy (sulfamethoxazole 400 mg/
trimethoprim 80 mg once or twice daily). Patients receiving antifungal
prophylaxis or preexisting antifungal treatment were excluded. Also,
AML with antecedent hematologic malignancies like Myelodysplastic
syndrome, and Myeloproliferative neoplasms, AML M3, relapsed AML
patients and chemotherapy courses with low-intensity regimen were
excluded. Baseline demographic and clinical data, type of AML,
chemotherapy courses, duration of febrile neutropenia, complete blood
cell count, cytogenetic risks, radiological examination"
high-resolution chest computed tomography (CT)", IFI incidence, site of
fungal infection, and patients outcome were recorded. Twenty age and
sex-matched healthy individuals were the control group in this study.
Diagnosis of Invasive Fungal Infection.
Diagnosis of IFI was applied according to 2008 consensus criteria of
the European Organization for Research and Treatment of Cancer/Invasive
Fungal Infections Cooperative Group (EORTC) and the National Institute
of Allergy and Infectious Diseases Mycoses Study Group (MSG), which
classified IFI into possible, probable, or proven IFI.[5]
Proven IFI requires that a fungus be detected by culture or
pathohistological blood analysis in a sterile site sample. Probable IFI
requires lesions on imaging indicative of fungal infection and
mycological evidence, not only culture and pathohistological analysis
of a sample but also indirect tests, such as galactomannan. Possible
IFI only requires imaging lesions indicating fungal infection without
presence mycological evidence.
Neutropenia was defined as a neutrophil count <500 cells/ μL.[19]
The duration of neutropenia in each course of chemotherapy was
collected. When patients remain febrile neutropenic >72 hrs after
antibacterial agent, a thorough history and physical examination were
recorded, along with culture for blood and other potentially infectious
focuses including oral mucositis grade ΙΙΙ or ΙѴ, or lower respiratory
tract infection (LRTI). For patients with no identified focuses, high
resolution computed tomography (CT) was performed, together with
galactomannan (GM) assay and PCR. Broncho-alveolar lavage (BAL) was not
performed routinely. Fluconazole 400 mg IV/day was given if IFI were
suspected with the CT findings, positive galactomannan or PCR assays,
or other clinical evidence.
Sample Collection and Processing.
Blood, oral swabs, and sputum samples were collected from the patients
according to their clinical presentation and different localizing signs
and symptoms before the initiation of antifungal therapy.
Identification of Candida spp.
Blood cultures were done by adding 5-10 mL blood to 50-100 mL Sabouraud
dextrose broth (Himedia, India) and incubated at 37ºC for 10 days with
subculture every other day.[20] Oral swabs and sputum
samples were cultivated on Sabouraud dextrose agar (Himedia, India)
with chloramphenicol (16 mg/mL). The isolates were further identified
by colony morphology on CHROMagar® Candida medium (CHROMagar, Paris,
France), germ tube test, chlamydospores on Tween 80 cornmeal agar
(Difco) and growth at 45°C.[21]
Identification of Mold. Direct
microscopic examination is performed on a fresh sample between a glass
slide and coverslip. The morphological characteristics of Aspergillus
spp are the presence of hyphae (hyaline and septate) with dichotomous
branches at 45° angles and with uniform width (3–6 µm). However, it is
hard to distinguish the species of Aspergillus because of the
difficulty in distinguishing the morphology of the different fungi
species. Aspergillus spp were
cultivated on Sabouraud’s dextrose-agar at 37 °C for 2 to 5 days. Fungi
that grew in culture were identified according to morphological and
microscopic criteria and Roth’s flag technique.[22,23]
In addition, Patient sera were tested for galactomannan (GM) by
Galactomannan ELISA kits according to the manufacturer instructions[24] (Bio-Rad, Hercules, CA). The presence of bacterial infections was tested by VITEK® 2 system.
DNA Extraction and PCR Amplification.
DNA extraction was performed using a commercial kit (QIAamp DNA Mini
Kit (Qiagen, Germany)) according to the manufacturer’s
instructions. PCR was performed utilizing the fungus specific,
universal primer pair ITS1 (ʹ5TCCGTAGGTGAACCTGCGG3ʹ) which hybridizes
at the 3ʹ end of 18S rDNA and ITS4 (ʹ5TCC TCC GCTTATGATAT GC3ʹ) which
hybridizes at the 5ʹend of 28S rDNA (Sigma, USA).[25]
The concentration was measured by a NanoDrop ND-1000 spectrophotometer
(NanoDrop Technologies). The PCR reaction mix contained 0.5 μM of each
primer, 10 μM deoxynucleotides, 1.5 mM MgCl2 and 1 x buffer (Promega).
One unit of the Taq Polymerase (Promega) was added to each tube. DNA
amplification was carried out in a Gene Amp9600 thermal cycler under
the following conditions: 35 cycles of denaturing at 94°C for 1 min;
annealing at 55.5°C for 2 min and extension at 72°C for 2 min; and
final extension at 72° for 10 min.[25,26] PCR products were visualized by electrophoresis on a 1% agarose gel stained with ethidium bromide.
Flow Cytometry.
Whole blood samples (anticoagulated with EDTA) were collected from
enrolled individuals and stained with the following antibodies (all
from BD bioscience, USA); Alexa fluor 488-conjugated anti-CD282 for
detection of TLR2, PE-conjugated anti-CD284|MD-2 complex for detection
of TLR4, PerCP-conjugated anti-HLA-DR, and APC-conjugated anti-CD3.
RBCs were lysed with the lysis buffer, then at least 10.000 events were
acquired and analyzed by FACS Caliber flow cytometer (BD bioscience,
USA). Appropriate isotype-matched controls were included in the
experiments to identify positive populations.[27] Data was analyzed with cell Quest software (BD bioscience, USA), (Figure 1).
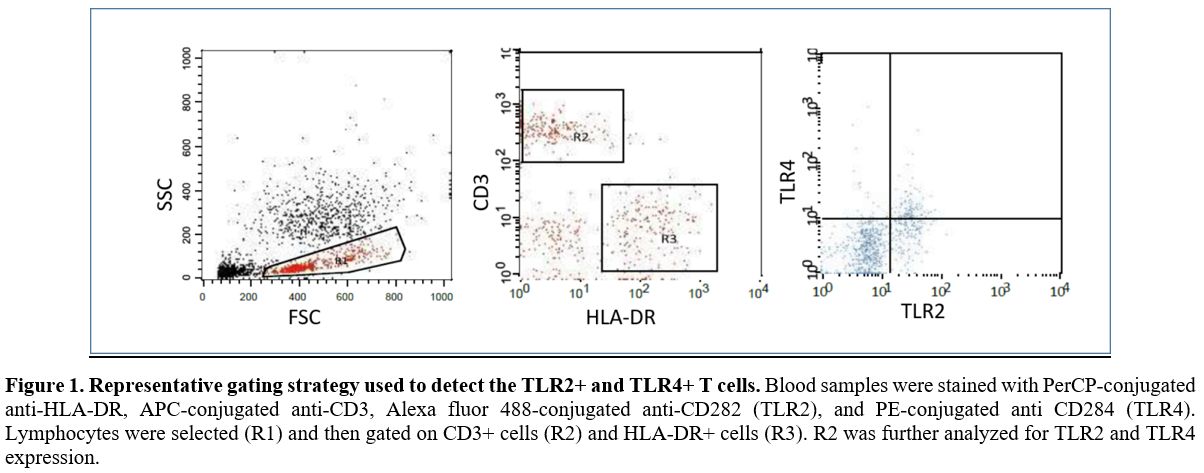 |
Figure
1. Representative gating strategy used to detect the TLR2+ and TLR4+ T cells.
Blood samples were stained with PerCP-conjugated anti-HLA-DR,
APC-conjugated anti-CD3, Alexa fluor 488-conjugated anti-CD282 (TLR2),
and PE-conjugated anti CD284 (TLR4). Lymphocytes were selected (R1) and
then gated on CD3+ cells (R2) and HLA-DR+ cells (R3). R2 was further
analyzed for TLR2 and TLR4 expression. |
Statistical Data.
Descriptive results of continuous variables were expressed as mean±SE
for non-parametric variables and as mean±SD for parametric variables.
Comparison of the demographic characteristics between cases and control
was calculated using the chi-square test for categorical data and
independent sample t-test for numerical variables. Qualitative
variables were expressed as the number of positive cases (%).
Differences in mean values of TLR2 and TLR4 level of expression between
different groups were calculated using the Mann-Whitney test. P-value
was considered significant at ˂ 0.05. Statistical calculation was
performed with the statistical package for social science software
(SPSS version 16.0 Inc, Chicago, III).
Results
From
2017 to 2020, 122 newly diagnosed non-M3 AML patients (aged more than
18) who received induction chemotherapy were admitted to Clinical
Hematology Unit, Internal Medicine Department, Assiut University
Hospital, South Egypt Cancer Institute (SECI). Forty patients (32.78%)
developed IFIs. The demographic and clinical characteristics of these
40 non-M3 AML patients with IFIs were presented in Table 1. The median
age was 38.8 years (range, 18–65 years); male patients were 27 (67.5%).
The diagnosis was applied according to the WHO criteria for AML. There
were mainly AML4, AML1 and AML2 (30 %, 25% and 22.5% respectively).
Eleven (27.5%) patients were favorable-cytogenetic group, 9 (22.5%)
poor group, and 20 (50%) the intermediate- risk group. No significant
differences were found between TLR4 or TLR2 expressions and age, sex,
Type of AML, cytogenetic risk and Patient’s outcome (P > 0.05).
The
most common sites of infection were the lower respiratory tract 47.5%
(19/40), and oral mucosa (mucositis grade ӀӀ or ӀѴ) 37.5% (15/40).
Mixed infection sites (bloodstream, oral, and LRTI) were detected only
in 15% (6/40), Table 1.
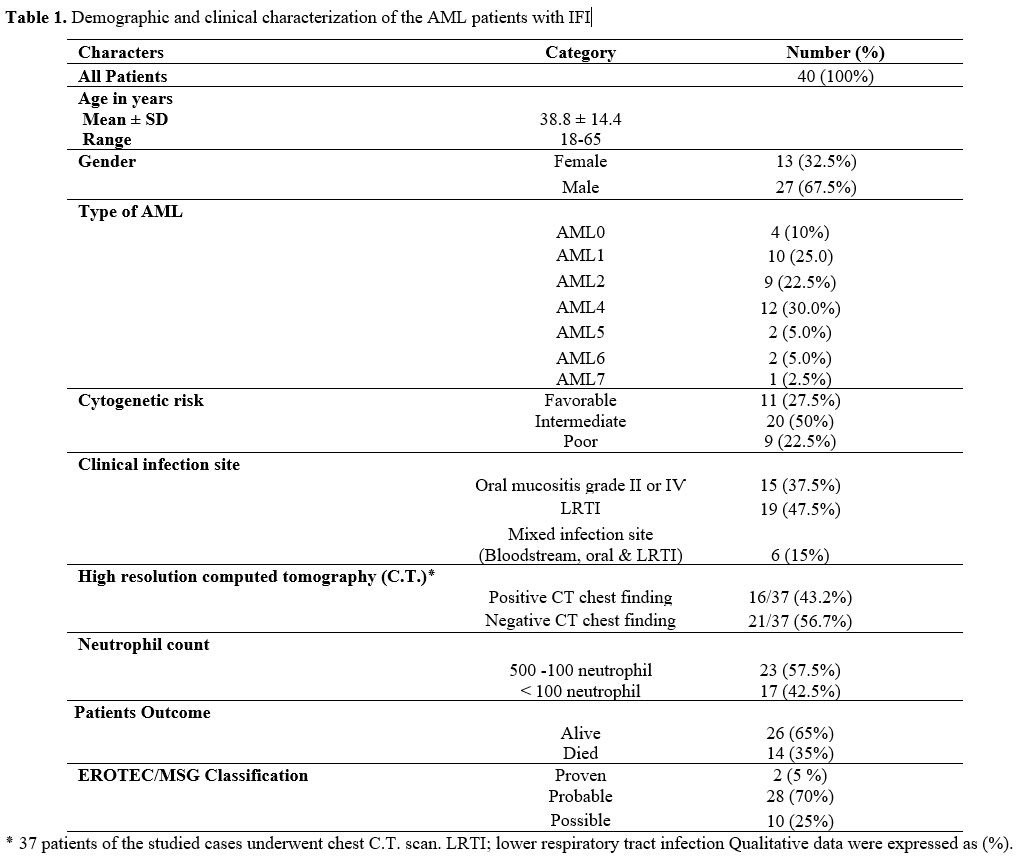 |
Table
1. Demographic and clinical characterization of the AML patients with IFI |
The
fungal pathogens among the 40 AML patients was identified as 2 (5%)
proven, 28 (70%) probable, and 10 (25%) possible IFIs. The pure fungal
growth was observed in 24 patients, whereas mixed bacterial and fungal
growth was encountered in 16 patients. Candida species was the most encountered fungi. It was present in 21 specimens (2 specimens were mixed candida and mold pathogen) followed by Aspargillus in 19 specimens then penicillum in 2 specimen, Table 2.
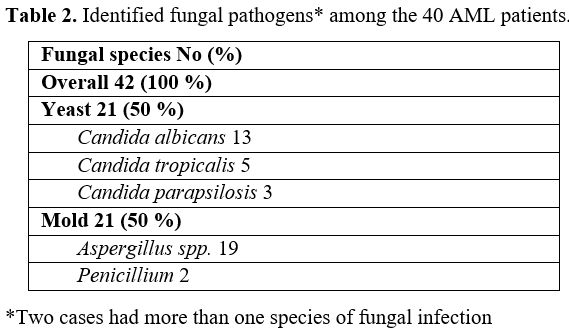 |
Table 2. Identified fungal pathogens* among the 40 AML patients. |
TLR2 expression in AML patients with IFIs in comparison to healthy controls showed no significant difference (p
꞊ 0.659), while there was a significant increase in the expression of
TLR4 in AML patients with IFI compared to healthy controls (p
= 0.001). TLR2 and TLR4 expression in AML patients with no IFI in
comparison to healthy controls had no significant difference (p ꞊ 0.72, 0.69 respectively), Table 3.
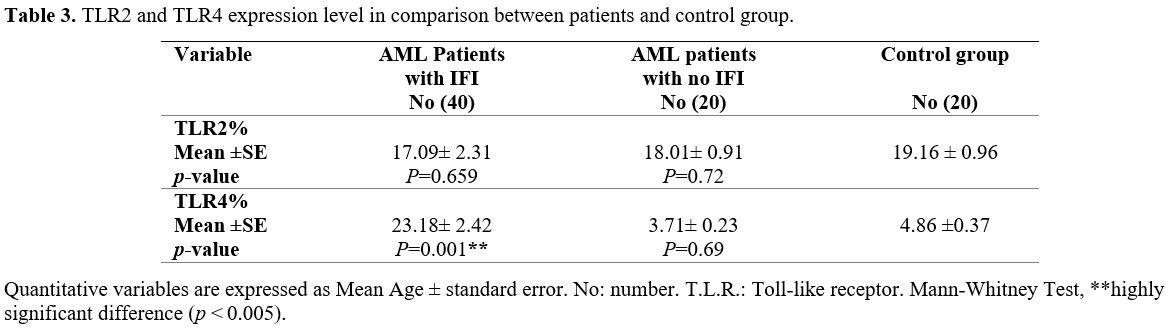 |
Table 3. TLR2 and TLR4 expression level in comparison between patients and control group. |
Moreover,
we observed that TLR2 expression increased significantly in AML
patients with mixed fungal and bacterial infections compared to healthy
controls (p = 0.002). Also, TLR4 expression in AML patients with mixed fungal and bacterial infection was significantly increased (p = 0.001), Table 4.
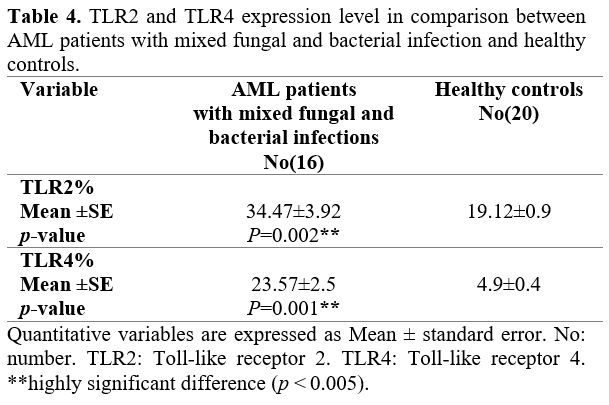 |
Table 4. TLR2 and
TLR4 expression level in comparison between AML patients with mixed
fungal and bacterial infection and healthy controls. |
Discussion
This
is the first study about T-lymphocytes expressions of TLRs and the
development of IFIs in AML patients receiving induction chemotherapy in
Assiut University Hospitals, and up to our knowledge in Egypt. We
reported that the overall incidence of IFIs in AML patients is 40/122
(32.78%), this incidence is considered high in comparison with other
reports from different countries.[28-30] In these
reports, the incidences of IFIs in AML patients varied from 4.0% to
48.4%. This variation is due to differences in patient populations,
chemotherapy regimens, antifungal prophylaxis, and geographic
variation. Recent study reported that (29%) of AML patients developed
an IFI. Patients with AML remain at risk for IFI despite the use of
several different antifungal agents for prophylaxis.[31]
The
high incidence in our study can be explained by many factors, including
limited health resources, the lack of routinely administered
anti-fungal prophylaxis, and environmental factors such as high
temperature, which facilitates fungal growth. This high incidence of
IFIs should start a new cost-effectiveness consideration about the
requirement of anti-fungal prophylaxis in AML patients with induction
chemotherapy. Hagiwara et al.[32] reported that AML
in developing countries with limited health resources, favors the
health authorities to use their low budget preferentially in another
illness that has a higher incidence and a better chance for achievement
of higher social impact.
The fungal pathogens among the 40 AML
patients was identified as 2 (5%) proven, 28 (70%) probable, and 10
(25%) possible IFIs. Tang et al.[33] reported that
the incidence of all-category IFIs was 34.6% (5.7% proven IFIs, 5.0%
probable IFIs and 23.8% possible IFIs). Nucci et al.[34] report a Brazilian incidence of (18.7%) for proven/probable IFIs in AML patients after diagnosis. Kim et al.[19] reported (9.6%) with 20 IFI diagnosed following HMA (three proven, four probable, 13 possible).
In
our study the pure fungal growth was observed in 24 (60%) patients,
whereas mixed bacterial and fungal growth was encountered in 16 (40%)
patients. Candida species was the most encountered fungi. It was present in 21 (50%) specimens (including 2 specimens were mixed candida and mold pathogen) followed by Aspargillus in 19 (45.2%) specimens then penicillum in 2 (4.8%) specimens. This result was different from Tang et al.[33] who reported that Candida spp still predominated and almost twice as common as Aspergillus spp.
The reasons for this difference are mostly due to difference in number
of patients enrolled, different specimen types and the absence of
anti-yeast azole prophylaxis.
The reports in Egypt are very
limited; an Egyptian study conducted on high-risk pediatric cancer
patients by EL-mahallawy et al.[35] reported that yeast was isolated in
(78.6%) of specimen and molds in (21.11%). Among yeasts, Candida was the commonest, while the most encountered molds were Aspergillus spp.
They found that polymicrobial (mixed bacterial and fungal growth) was
encountered in 62.5% of specimen, which is in great accordance with our
results.
In this study, non-albicans Candida spp. (C. tropicalis and C. parapsilosis) were common 8/21 (38.1%) as C. albicans 13/21 (61.9%). Another study with similar findings postulated that neutropenia is correlated with non-albican Candida infections.[36]
An Egyptian study reported that 75 (44.1%) Candida spp (25 (33.3%) non-albicans Candida spp and 50 (66.6%) C. albicans) were isolated from AML patients on induction chemotherapy.[37]
The common site of IFI was the lower respiratory tracts (47.5%, 19/40),
and oral mucosa (mucositis grade ӀӀ or ӀѴ) (37.5%, 15/40) followed by
mixed infection sites (bloodstream, oral and LRTI) (15%, 6/40). Pagano
et al.[38] and Tang et al.[33]
reported that lower respiratory tract was the most common site for IFIs
(80% and 75.4% respectively); also EL-mahallawy et al.[35] and Kurosawa et al.[30]
found an incidence of (35.7% and 55.3% respectively) for IFIs affecting
the lung. Few articles evaluated the risk factors of IFIs in AML
patients during induction chemotherapy. In this study, we have
determined these risk factors as standard induction chemotherapy,
febrile neutropenia, elderly and male gender. Tang et al.[35]
postulated similar risk factors including standard induction
chemotherapy, younger than 40 or older than 60 years, and a poor
chemotherapy response for all-category IFIs. Neofytos et al.[39]
postulated that mucositis and organ dysfunctions are important risk
factors for invasive candidiasis during induction chemotherapy, and
male gender is the only risk factor for mold infection. Hammond et al.[29] also reported male gender as risk factors for IFIs. Chen et al.[40]
stated that AML patients have multiple risk factors for developing
invasive fungal diseases, such a including advanced age, prolonged and
profound neutropenia, the presence of indwelling catheters, and
individual genetic susceptibilities. Previous results indicate the
heterogeneity of the study subjects and treatment protocols.
The
exact role of TLRs in the development of invasive fungal infection in
AML patients is unknown. Numerous endogenous and exogenous factors
affect cell proliferation and play critical roles in cancer
development. The expression level of TLRs may depend on the
environment, subset, cell type, stimulus and probably age group.[41]
In
the current study, no statistically significant differences were found
between TLR4 or TLR2 expressions and age, sex, cytogenetic risk and
Patient’s outcome (P > 0.05). Similar results showed by Ramzi et al.[42] postulated that expressions of TLR2 did not show significant differences in cytogenetic abnormalities status (P = 0.67). The expression of TLR4 was not different in favorable, intermediate and poor risk groups (P = 0.97). Renshaw et al[41]
reported that old age could have negative effects on TLR expression and
function, and therefore leads to increased susceptibility to infections
and poor adaptive immune responses.
The current study
included 122 newly diagnosed non-M3 AML patients and reporting no
statistically significant differences between TLR4 or TLR2 expressions
and type of AML (P > 0.05). In the same context, Ramzi et al.[42] observe a higher expression of TLR2 in AML-M3 cases compared to non-M3 AML patients (P = 0.015).
Human
T cells isolated from peripheral blood reported to express mRNA for
most TLRs, with considerable variation in the reported expression
levels. Protein expression of TLR2, 3, 4, 5 and 9 has also been
detected by flow cytometry.[43] The current study
revealed that TLR2 expression in AML patients with IFIs in comparison
to healthy controls presented no significant difference (p
꞊ 0.659), while there was a significant increase in the expression of
TLR4 in the same patients group compared to healthy controls (p
= 0.001). Consistent with these findings is the study of
Bellocchio, Montagnoli. They reported that TLR4 but not TLR2
participated in host defense against A. fumigatus.[44] In addition, Chai et al.[45] stated that after stimulation with A. fumigatus
conidia, surface TLR2 expression is markedly reduced compared to TLR4
expression, this suggests that A. fumigatus conidia induced depletion
and downregulation of the TLR2-mediated pathway involved in the
receptor internalization together with Aspergillus conidia into the phagosome, resulting in decreased TLR2 expression on the cell membrane. Chai et al [45]
suggested a possible explanation for these findings as they postulated
that the balance between TH1 and TH2 immune system pathways is
necessary for the pathogen clearance and limitation of
inflammation. TLR4 favors the production of TH1 response with
pro-inflammatory cytokine production such as IFN-γ and 1L- 12, which
induces protective antifungal defense mechanisms. T regulatory cells
induced by TH2 response mediated by TLR2 signaling are needed to lower
immune response and to avoid collateral damage after antifungal TH1
response mediated by TLR4 signaling.
Our result revealed that
TLR2 and TLR4 expression in patients with polymicrobial infection
(fungus and bacteria) are significantly increased as compared to
healthy controls. This result agreed with the result of Armstrong
et al.[46] who reported that expression of TLR2 and
TLR4 in septic patients was significantly up-regulated compared with
the expression of these receptors in healthy individuals. Tsujimoto et
al.[47] stated also that septic patients display significantly up-regulated TLR expression in various organs.
We
can conclude that in polymicrobial infection (fungus and bacteria)
there is a marked increase of both TLR2 and TLR4 expression and this
may be due to the powerful effect of bacterial LPS and other bacterial
PAMPs that augment the stimulatory effect of fungal PAMPs.
Susceptibility
to infections is determined by the malignant disease and its treatment,
environmental factors (e.g. nutritional status and hygiene of the
patient), and genetically determined variations of the immune system.
Some genetic polymorphisms in the innate immune system, such as
profound mannose-binding lectin deficiency and TLR polymorphism
associated with an increased risk of infections. Mutations in genes
encoding TLRs or downstream signaling proteins increase the risk of
infection.[48]
Numerous polymorphisms and mutational inactivation have been described in TLRs and appear to have clinical significance,[48,47] reported
that severely septic patients with bad general conditions and the
unfavorable clinical outcome did not have increased expression of TLRs,[49] have
observed that a decrease in TLR2 expression in patients with invasive
candidiasis can lead to severe disseminated infection. On the
other side,[50] found that mice with non-functional TLR4 showed increased fungal load in the kidneys and deficiencies in neutrophil upon C. albicans challenge when compared to TLR4 responsive mice.
Conclusions
The
incidence of IFIs is high in AML patients who received induction
chemotherapy in Assiut University Hospitals. TLR2 and TLR4
expressions in AML patients with IFI are related to invasiveness and
dissemination of fungal infection. TLRs expressions could be important
biological markers for the occurrence of IFI in non-M3 AML patients
after intensive induction chemotherapy. Additional larger studies
including a larger number of patients and detection of proinflammatory
cytokines are necessary to confirm the immunological relation between
TLR and fungal infection in AML patients.
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
M.R.A.
and D.S. conceived and designed the research. D.S., M.R.A. and S.M.M.
recruited patients, carried out the clinical investigations, collected
clinical data. S.G.E. and M.A.E. contributed in the interpretation of
data for the work. D.S., M.R.A., S.M.M., S.G.E. and M.A.E. prepared the
original draft of the manuscript. All authors contributed to data
analysis, drafting or revising the article, have agreed on the journal
to which the article will be submitted, gave final approval of the
version to be published, and agree to be accountable for all aspects of
the work.
References
- Wang ES. Common fungal infections in patients with leukemia. Clin Adv Hematol Oncol. 2017;15(5):352-4.
- Heimann
SM, Vehreschild MJ, Cornely OA, Franke B, von Bergwelt-Baildon M,
Wisplinghoff H, et al. A cost and resource utilization analysis of
micafungin bridging for hemato-oncological high-risk patients
undergoing allogeneic stem cell transplantation. Eur J Haematol.
2015;94(6):526-31. https://doi.org/10.1111/ejh.12466 PMid:25310918
- Rambaldi
B, Russo D, Pagano L. Defining Invasive Fungal Infection Risk in
Hematological Malignancies: A New Tool for Clinical Practice. Mediterr
J Hematol Infect Dis. 2017;9(1):e2017012. https://doi.org/10.4084/mjhid.2017.012 PMid:28101316 PMCid:PMC5224802
- Heinz
WJ, Buchheidt D, Christopeit M, von Lilienfeld-Toal M, Cornely OA,
Einsele H, et al. Diagnosis and empirical treatment of fever of unknown
origin (FUO) in adult neutropenic patients: guidelines of the
Infectious Diseases Working Party (AGIHO) of the German Society of
Hematology and Medical Oncology (DGHO). Ann Hematol.
2017;96(11):1775-92. https://doi.org/10.1007/s00277-017-3098-3 PMid:28856437 PMCid:PMC5645428
- DePauw
B, Walsh TJ, Donnelly JP, Stevens DA, Edwards JE, Calandra T, et al.
Revised definitions of invasive fungal disease from the European
Organization for Research and Treatment of Cancer/Invasive Fungal
Infections Cooperative Group and the National Institute of Allergy and
Infectious Diseases Mycoses Study Group (EORTC/MSG) C. Clin Infect Dis.
2008;46(12): 1813-21. https://doi.org/10.1086/588660 PMid:18462102 PMCid:PMC2671227
- J
Peter Donnelly, Sharon C Chen, Carol A Kauffman, William J Steinbach,
John W Baddley, Paul E Verweij, Cornelius J Clancy et al. Revision and
Update of the Consensus Definitions of Invasive Fungal Disease From the
European Organization for Research and Treatment of Cancer and the
Mycoses Study Group Education and Research Consortium, Clinical
Infectious Diseases, 2020; 71( 6)15: 1367-1376.
- Ferwerda
G, Netea MG, Joosten LA, van der Meer JW, Romani L, Kullberg BJ. The
role of Toll-like receptors and C-type lectins for vaccination against
Candida albicans. Vaccine. 2010 Jan 8;28(3):614-22. doi:
10.1016/j.vaccine.2009.10.082. Epub 2009 Nov 1. PMid:19887129
- Goyal
S, Castrillón-Betancur JC, Klaile E, Slevogt H. The Interaction of
Human Pathogenic Fungi With C-Type Lectin Receptors. Front Immunol.
2018 Jun 4;9:1261. doi: 10.3389/fimmu.2018.01261. https://doi.org/10.3389/fimmu.2018.01261 PMid:29915598 PMCid:PMC5994417
- McGettrick
AF, O'Neill LA. Tolllike receptors: key activators of leucocytes and
regulator of haematopoiesis. Br JHaematol (2007) 139:185. doi:
10.1111/j.1365-2141.2007.06802.x https://doi.org/10.1111/j.1365-2141.2007.06802.x PMid:17897294
- Zanin-Zhorov
A and Cohen IR (2013) Signaling via TLR2 and TLR4 directly
down-regulates T cell effector functions: the regulatory face of danger
signals. Front. Immunol. 4:211. doi: 10.3389/fimmu.2013.00211 https://doi.org/10.3389/fimmu.2013.00211 PMid:23898332 PMCid:PMC3722573
- Gururajan
M, Jacob J, Pulendran B. Toll-like receptor expression and
responsiveness of distinct murine splenic and mucosal B-cell subsets.
PLoS ONE (2007) 2:e863. doi:10. 1371/journal.pone.0000863 https://doi.org/10.1371/journal.pone.0000863 PMid:17848994 PMCid:PMC1955832
- Zanin-Zhorov
A, Nussbaum G, Franitza S, Cohen IR, Lider O. Tcells
respondtoheatshockprotein60via TLR2: activation of adhesion and
inhibition of chemokine receptors. FASEB J (2003) 17:1567. https://doi.org/10.1096/fj.02-1139fje PMid:12824285
- Zanin-Zhorov
A, Cahalon L, Tal G, Margalit R, Lider O, Cohen IR. Heat shock protein
60 enhances CD4+ CD25+ regulatory T cell function via innate TLR2
signaling. J Clin Invest (2006) 116:2022. doi: 10.1172/JCI28423 https://doi.org/10.1172/JCI28423 PMid:16767222 PMCid:PMC1474819
- Tomita
T, Kanai T, Fujii T, Nemoto Y, Okamoto R, Tsuchiya K, et al.
MyD88-dependent pathway in T cells directly modulates the expansion of
colitogenic CD4+ T cells in chronic colitis. J Immunol. 2008; 180:5291.
https://doi.org/10.4049/jimmunol.180.8.5291 PMid:18390710
- Cottalorda
A, Verschelde C, Marcais A, Tomkowiak M, Musette P, Uematsu S, et al.
TLR2 engagement on CD8 T cells lowers the threshold for optimal
antigen-induced T cell activation. Eur J Immunol. 2006; 36:1684. https://doi.org/10.1002/eji.200636181 PMid:16761317
- Gelman
AE, Zhang J, Choi Y, Turka LA. Toll-like receptor ligands directly
promote activated CD4+ T cell survival. J Immunol. 2004; 172:6065.
https://doi.org/10.4049/jimmunol.172.10.6065 PMid:15128790 PMCid:PMC2833313
- Netea
MG, Van der Meer JW, Kullberg BJ (2007). Recognition of fungal
pathogens by Toll-like receptors. In Immunology of Fungal Infections.
Eur J Clin Microbiol Infect Dis, 23, 672-6. https://doi.org/10.1007/1-4020-5492-0_11
- Sabattini
E, Bacci F, Sagramoso C, Pileri SA. WHO classification of tumours of
haematopoietic and lymphoid tissues in 2008: an overview. Pathologica.
2010;102(3):83-7.
- Kim GYG, Burns J,
Freyer CW, Hamilton KW, Frey NV, Gill SI, et al. Risk of invasive
fungal infections in patients with high-risk MDS and AML receiving
hypomethylating agents. Am J Hematol. 2020;95(7):792-8. https://doi.org/10.1002/ajh.25808 PMid:32242967
- Power D, Johnson J. Difco™ and BBL™ manual: manual of microbiological culture media. Becton Dickinson and Company, Sparks; 2009.
- Pincus DH, Orenga S, Chatellier S. Yeast identification--past, present, and future methods. Med Mycol. 2007;45(2):97-121. https://doi.org/10.1080/13693780601059936 PMid:17365647
- Fromtling
R, Rhodes J, Dixon D. Taxonomy, classification, and morphology of the
Fungi. J Manual of clinical microbiology. 2003;2:1653-8.
- Desoubeaux
G, Bailly E, Chandenier J. Diagnosis of invasive pulmonary
aspergillosis: updates and recommendations. Med Mal Infect.
2014;44(3):89-101. https://doi.org/10.1016/j.medmal.2013.11.006 PMid:24548415
- Ascioglu
S, Rex JH, de Pauw B, Bennett JE, Bille J, Crokaert F, et al. Defining
opportunistic invasive fungal infections in immunocompromised patients
with cancer and hematopoietic stem cell transplants: an international
consensus. Clin Infect Dis. 2002;34(1):7-14. https://doi.org/10.1086/323335 PMid:11731939
- Karakousis
A, Tan L, Ellis D, Alexiou H, Wormald PJ. An assessment of the
efficiency of fungal DNA extraction methods for maximizing the
detection of medically important fungi using PCR. J Microbiol Methods.
2006;65(1):38-48 https://doi.org/10.1016/j.mimet.2005.06.008 PMid:16099520
- Williamson
EC, Leeming JP, Palmer HM, et al. Diagnosis of invasive aspergillosis
in bone marrow transplant recipients by polymerase chain reaction. Br.
J. Haematol. 2000;108 (1):132-139. https://doi.org/10.1046/j.1365-2141.2000.01795.x PMid:10651736
- Kurt-Jones
EA, Mandell L, Whitney C, Padgett A, Gosselin K, Newburger PE, et al.
Role of toll-like receptor 2 (TLR2) in neutrophil activation: GM-CSF
enhances TLR2 expression and TLR2-mediated interleukin 8 responses in
neutrophils. Blood. 2002;100(5):1860-8. https://doi.org/10.1182/blood.V100.5.1860.h81702001860_1860_1868 PMid:12176910
- Gomes
MZ, Mulanovich VE, Jiang Y, Lewis RE, Kontoyiannis DP. Incidence
density of invasive fungal infections during primary antifungal
prophylaxis in newly diagnosed acute myeloid leukemia patients in a
tertiary cancer center, 2009 to 2011. Antimicrob Agents Chemother.
2014;58(2):865-73. https://doi.org/10.1128/AAC.01525-13 PMid:24277033 PMCid:PMC3910838
- Hammond
SP, Marty FM, Bryar JM, DeAngelo DJ, Baden LR. Invasive fungal disease
in patients treated for newly diagnosed acute leukemia. Am J Hematol.
2010;85(9):695-9. https://doi.org/10.1002/ajh.21776 PMid:20652970
- Kurosawa
M, Yonezumi M, Hashino S, Tanaka J, Nishio M, Kaneda M, et al.
Epidemiology and treatment outcome of invasive fungal infections in
patients with hematological malignancies. Int J Hematol.
2012;96(6):748-57. https://doi.org/10.1007/s12185-012-1210-y PMid:23111539
- Wasylyshyn
A, Linder KA, Castillo CG, Zhou S, Kauffman CA, Miceli MH. Breakthrough
Invasive Fungal Infections in Patients with Acute Myeloid Leukemia.
Mycopathologia. 2020;185(2):299-306. https://doi.org/10.1007/s11046-019-00418-8 PMid:31939052
- Hagiwara
M, Sharma A, Chung KC, Delea TE. Burden of acute myeloid leukemia (AML)
in a US commercially insured population. American Society of Clinical
Oncology; 2017. https://doi.org/10.1200/JCO.2017.35.15_suppl.e18330
- Tang
JL, Kung HC, Lei WC, Yao M, Wu UI, Hsu SC, et al. High Incidences of
Invasive Fungal Infections in Acute Myeloid Leukemia Patients Receiving
Induction Chemotherapy without Systemic Antifungal Prophylaxis: A
Prospective Observational Study in Taiwan. PLoS One.
2015;10(6):e0128410. https://doi.org/10.1371/journal.pone.0128410 PMid:26061179 PMCid:PMC4462587
- Nucci
M, Garnica M, Gloria AB, Lehugeur DS, Dias VC, Palma LC, et al.
Invasive fungal diseases in haematopoietic cell transplant recipients
and in patients with acute myeloid leukaemia or myelodysplasia in
Brazil. Clin Microbiol Infect. 2013;19(8):745-51. https://doi.org/10.1111/1469-0691.12002 PMid:23009319
- El-Mahallawy
HA, Shaker HH, Ali Helmy H, Mostafa T, Razak Abo-Sedah A. Evaluation of
pan-fungal PCR assay and Aspergillus antigen detection in the diagnosis
of invasive fungal infections in high-risk paediatric cancer patients.
Med Mycol. 2006; 44(8):733-9. https://doi.org/10.1080/13693780600939955 PMid:17127630
- Chi
HW, Yang YS, Shang ST, Chen KH, Yeh KM, Chang FY, et al. Candida
albicans versus non-albicans bloodstream infections: the comparison of
risk factors and outcome. J Microbiol Immunol Infect.
2011;44(5):369-75. https://doi.org/10.1016/j.jmii.2010.08.010 PMid:21524971
- Sayed
SA, Hassan EAB, Abdel Hameed MR, Agban MN, Mohammed Saleh MF, Mohammed
HH, Abdel-Aal ABM, Elgendy SG. Ketorolac-fluconazole: A New Combination
Reverting Resistance in Candida albicans from Acute Myeloid Leukemia
Patients on Induction Chemotherapy: In vitro Study. J Blood Med.
2021;12:465-474. https://doi.org/10.2147/JBM.S302158 PMid:34163275 PMCid:PMC8214543
- Pagano
L, Girmenia C, Mele L, Ricci P, Tosti ME, Nosari A, et al. Infections
caused by filamentous fungi in patients with hematologic malignancies.
A report of 391 cases by GIMEMA Infection Program. Haematologica.
2001;86(8):862-70.
- Neofytos D, Lu K,
Hatfield-Seung A, Blackford A, Marr KA, Treadway S, et al.
Epidemiology, outcomes, and risk factors of invasive fungal infections
in adult patients with acute myelogenous leukemia after induction
chemotherapy. Diagn Microbiol Infect Dis. 2013;75(2):144-9. https://doi.org/10.1016/j.diagmicrobio.2012.10.001 PMid:23142166 PMCid:PMC3986043
- Chen
MJ, HU Rong, Jiang XY, Wu Y, HE Z, Chen J, Zhan L.Dectin-1 rs3901533
and rs7309123 Polymorphisms Increase Susceptibility to Pulmonary
Invasive Fungal Disease in Patients with Acute Myeloid Leukemia from a
Chinese Han Population. Current Medical Science, 39(6):906-912,2019. https://doi.org/10.1007/s11596-019-2122-3 PMid:31845221
- Renshaw
M, Rockwell J, Engleman C, et al. Cutting edge: impaired Toll-like
receptor expression and function in aging. J Immunol.
2002;169(9):4697-701. https://doi.org/10.4049/jimmunol.169.9.4697 PMid:12391175
- Ramzi
M, Khalafi-Nezhad A, Iravani Saadi M, Jowkar Z. Association between
TLR2 and TLR4 Expression and Response to Induction Therapy in Acute
Myeloid Leukemia Patients. Int J Hematol Oncol Stem Cell Res. 2018 Oct
1;12(4):303-312. https://doi.org/10.18502/ijhoscr.v12i4.109 PMid:30774831 PMCid:PMC6375370
- Rahman
AH, Taylor DK, Turka LA. The contribution of direct TLR signaling to T
cell responses. Immunol Res. 2009;45(1):25-36. doi:
10.1007/s12026-009-8113-x. https://doi.org/10.1007/s12026-009-8113-x PMid:19597998 PMCid:PMC4486050
- Bellocchio
S, Montagnoli C, Bozza S, Gaziano R, Rossi G, Mambula SS, et al. The
contribution of the Toll-like/IL-1 receptor superfamily to innate and
adaptive immunity to fungal pathogens in vivo. J Immunol.
2004;172(5):3059-69. https://doi.org/10.4049/jimmunol.172.5.3059 PMid:14978111
- Chai
LY, Netea MG, Vonk AG, Kullberg BJ. Fungal strategies for overcoming
host innate immune response. Med Mycol. 2009;47(3):227-36. https://doi.org/10.1080/13693780802209082 PMid:18654922
- Armstrong
L, Medford AR, Hunter KJ, Uppington KM, Millar AB. Differential
expression of Toll-like receptor (TLR)-2 and TLR-4 on monocytes in
human sepsis. Clin Exp Immunol. 2004;136(2):312-9. https://doi.org/10.1111/j.1365-2249.2004.02433.x PMid:15086396 PMCid:PMC1809013
- Tsujimoto
H, Ono S, Efron PA, Scumpia PO, Moldawer LL, Mochizuki H. Role of
Toll-like receptors in the development of sepsis. Shock.
2008;29(3):315-21. https://doi.org/10.1097/SHK.0b013e318157ee55 PMid:18277854
- Pamer EG. TLR polymorphisms and the risk of invasive fungal infections. N Engl J Med. 2008;359(17):1836-8. https://doi.org/10.1056/NEJMe0806412 PMid:18946070 PMCid:PMC2630794
- Villamón
E, Gozalbo D, Roig P, O'Connor JE, Ferrandiz ML, Fradelizi D, et al.
Toll-like receptor 2 is dispensable for acquired host immune resistance
to Candida albicans in a murine model of disseminated candidiasis. J
Microbes. 2004;6(6):542-8. https://doi.org/10.1016/j.micinf.2004.02.015 PMid:15158187
- Netea
MG, Van Der Graaf CA, Vonk AG, Verschueren I, Van Der Meer JW, Kullberg
BJ. The role of toll-like receptor (TLR) 2 and TLR4 in the host defense
against disseminated candidiasis. J Infect Dis. 2002;185(10):1483-9. https://doi.org/10.1086/340511 PMid:11992285
[TOP]



