Association of Mycobacterium Tuberculosis Lineages with IFN-γ
and TNF-α Gene Polymorphisms among Pulmonary Tuberculosis Patient
Jayastu Senapati1,
Anup J. Devasia1, Abhijeet Ganapule1,
Leni George2 and Auro Viswabandya1
1
Department of Clinical Haematology, Christian Medical College and
Hospital, Vellore-632004. India.
2 Department of Dermatology, Christian Medical
College and Hospital, \Vellore-632004. India.
Corresponding author: Dr. Auro Viswabandya, MD, DM. Professor,
Department of Clinical
Haematology, Christian Medical College, Vellore – 632004. Tamil Nadu –
India. Tel. 0416-3072352/2282352, Fax. 0416 2226449/2232035. E-mail:
aurov@cmcvellore.ac.in
Published: February 17, 2014
Received: October 19, 2013
Accepted: January 15, 2014
Meditter J Hematol Infect Dis 2014, 6(1): e2014016, DOI
10.4084/MJHID.2014.016
This article is available on PDF format at:

This
is an Open Access article distributed
under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Sorafenib
is a novel small molecule multiple kinase inhibitor which has been used
for metastatic renal cancer, hepatocellular cancer. Sorafenib induced
skin rash has been discussed as a side effect in trials in both, FLT3
wild type and mutated acute myeloid leukemia (AML), as monotherapy or
as combination with other chemotherapeutic agents . We describe a
patient with FLT 3 ITD mutated AML, who was started on adjunctive
Sorafenib therapy. Skin reactions manifested as NCI Grade III
palmoplantar erythrodysesthesia (PPE), requiring drug discontinuation.
Several pathogenic mechanisms have been implicated in Sorafenib induced
skin reactions, but none has been conclusively proven. While treatment
options are varied for early stage skin reactions, drug discontinuation
remains the only possible therapy presently for severe grade skin
reaction.
|
Introduction
Sorafenib induced skin rash has been
widely described in context to its use in advanced renal and
hepatocellular cancer.[1–4]
Here we describe a case of a classical Sorafenib induced hand foot skin
rash (HFSR) in a patient with acute myeloid leukemia (AML).
Case Report
A 63 years old lady, with no significant history was diagnosed with AML
with myelodysplasia related changes. Her cytogenetic analysis revealed
normal karyotype and molecular analysis showed positivity for FLT3 ITD
and NPM 1 frame shift mutation. She was initially started on
Azacytidine based chemotherapy given her
age and poor
general condition. The family had been made aware of the poor outcome
and had opted for best supportive therapy with Azacytidine. After 2
cycles of the same as there was disease progression, she was started on
cytosine and daunorubicin chemotherapy (5+2) with Sorafenib as an
adjunct therapy at a dose of 400 mg twice daily. Ten days after
starting Sorafenib she complained of bilateral heel pain while walking,
associated with paresthesia and erythema of the skin which increased
over the next 2 days with formation of bulla and hyperesthesia. Similar
rashes were also noticed over her palms with discoloration of nail beds
(Figure 1).
She had difficulty in performing her activities of daily life. A
dermatology opinion was sought, and a diagnosis of Sorafenib induced
HFSR was made. She was staged as NCI Grade III/WHO Grade IV and was
started on topical tacrolimus and clobetasol along with analgesics. In
view of progressive skin manifestations and inadequate pain relief with
analgesics, Sorafenib was discontinued on day 12 of therapy. Three days
after stopping Sorafenib there was decrease in heel pain, and the
erythema decreased with healing of blisters. She became NCI grade II
within 3 days of stopping Sorafenib and Grade I within 6 days (Figure 2). She
became asymptomatic by another 7 days and was ambulant normally.
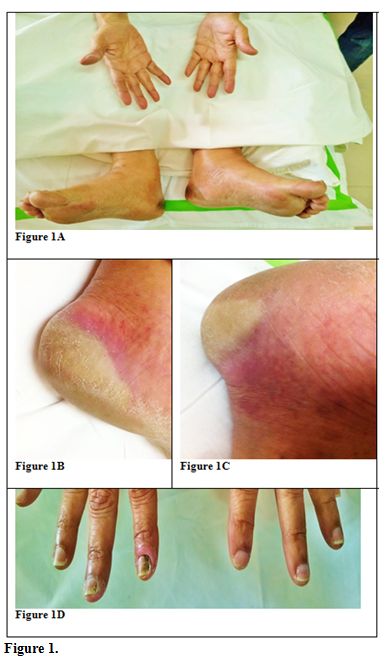 | Figure 1. |
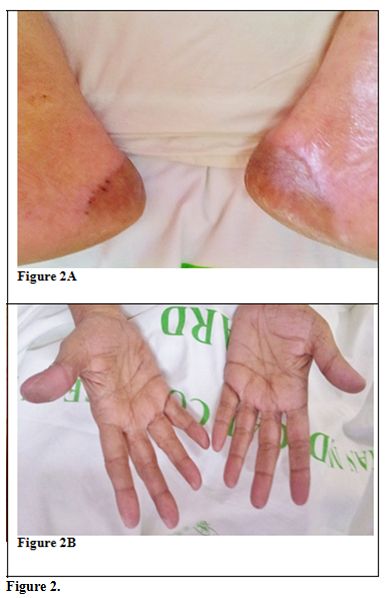 | Figure 2 |
Discussion
Sorafenib is a novel multiple tyrosine kinase inhibitor which has shown
efficacy in FLT3 ITD mutated AML, alone or in addition to other
cytotoxicity drugs.[5–11] We
discuss here the
important clinical features of HFSR and management algorithm. HFSR have
been described as the most common side effect with the use of Sorafenib
in solid tumours. A systematic analysis in solid tumours showed an
overall incidence of all grade Sorafenib HFSR of 33.8 % with 8.9% being
Grade III.[12] HFSR has been
described as one of the
commonest toxicities of Sorafenib; in several trials using Sorafenib as
a single agent or as add-on therapy to other cytotoxic therapies in
leukemia. Table 2
lists the
major trials and the frequencies of HFSR in study patients. This to our
knowledge is the first reported case, of Sorafenib induced HFSR,
outside trial data, in a case of acute myeloid leukemia where Sorafenib
was used as add on therapy along with standard chemotherapy.
HFSR associated with Sorafenib belongs to the spectrum of PPE, which is
associated with the use of several cytotoxic chemotherapy drugs, the
most common being capecitabine and 5-fluoro-uracil. Table 3
lists the common cytotoxic medications associated with PPE. However,
the HFSR associated with Sorafenib therapy focally affects the weight
and friction bearing acral surfaces, unlike the classic hand foot
syndrome (HFS) reported with other chemotherapeutic agents.[13] HFS usually presents as diffuse
erythema of palms and soles that does not have predilection to areas of
friction or trauma.[14]
Sorafenib induced HFSR can present in varied forms, ranging from mild
acral erythema to severe hyperesthesia with desquamation of the skin
leading to severe morbidity and drug discontinuation. This skin rash
has classically been graded by the National Cancer Institute (NCI) and
World Health Organisation (WHO) grading systems (Table 1).
Several mechanisms have been implicated in the pathogenesis of
Sorafenib induced skin rash. Direct cytotoxicity by increased
concentration of the drug in the palmo-plantar eccrine glands has been
postulated.[15] Inhibition of
vascular endothelial
growth factor (VEGF) and platelet derived growth factor receptors
(PDGFR) seem to play a role.[16]
Due to inhibition of
growth and repair pathways mediated by the above mentioned
pro-angiogenic receptors, areas subjected to high pressure and friction
are prone for HFSR with Sorafenib.[17]
VEGF seems to
play an important role as combination of Bevacizumab which is a
specific antibody against VEGF, with Sorafenib increases the incidence
of HFSR, while HFSR has not been described in Bevacizumab monotherapy.[13]
Therapy depends on the stage of HFSR. While early stage lesions require
only regular dermatological evaluation, emollients, adequate protection
from environment; advanced stage lesions require topical
immunomodulators, dose modification and often discontinuation of the
drug (Table 4).[16]
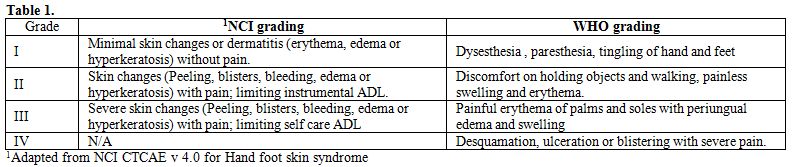 | Table 1. Demographic
of study populations |
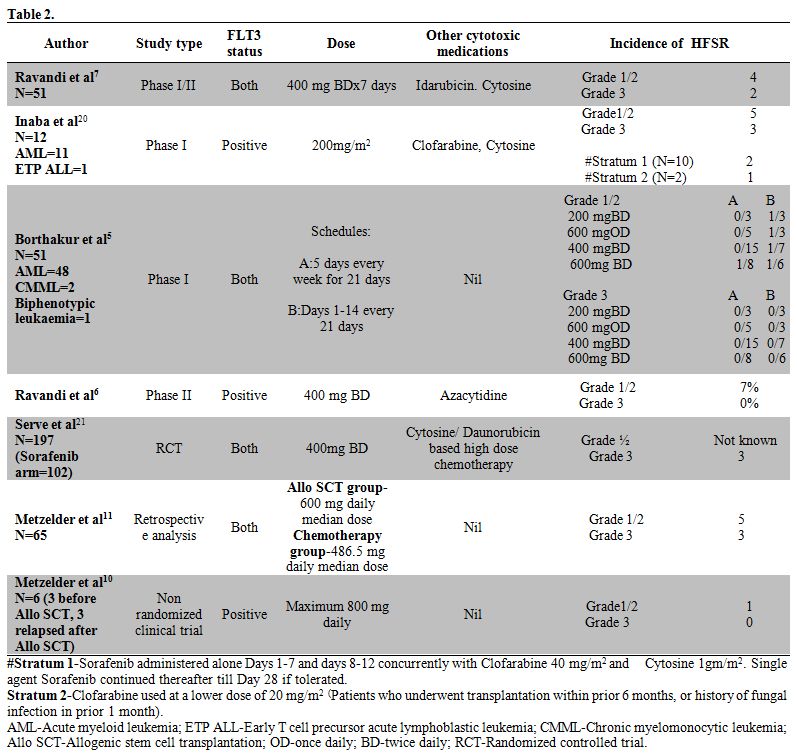 | Table 2. |
 |
Table 3. Common cytotoxic drugs associated with
Palmoplantar erythrodysesthesia |
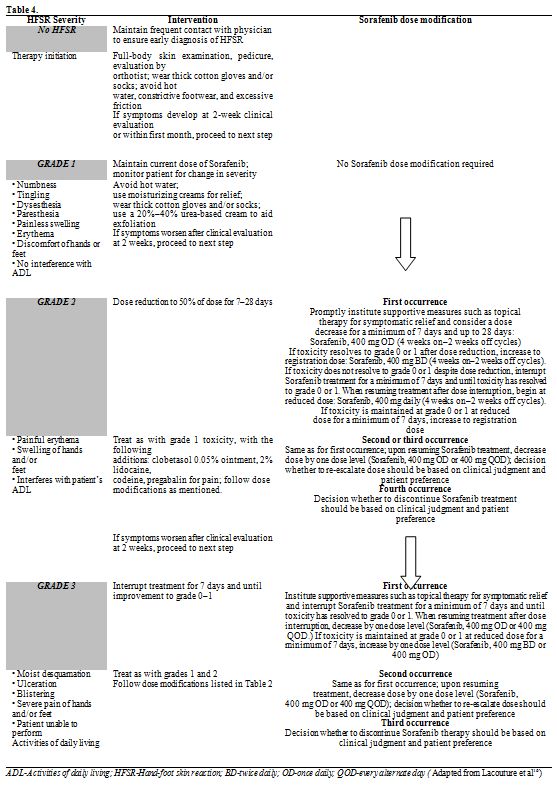 | Table 4. |
In
our patient, the progression of cutaneous symptoms from NCI Grade I to
NCI grade III over a span of 2 days, even with adequate local
immunomodulatory therapy and analgesics, required discontinuation of
the drug. There was a significant response to discontinuation of the
drug, and her symptoms downgraded to Grade I by 4 days. It is difficult
to say whether the daunorubicin and cytosine had any role in her
cutaneous symptoms, but looking at the distribution pattern of the skin
rash and the temporal profile of the Sorafenib administration and
discontinuation in conjunction with her cutaneous findings, it is
fairly clear that it is primarily due to Sorafenib.
Conclusions
Sorafenib induces HFSR less frequently in acute myeloid
leukemia
than in solid cancers treated together with Bevacizumab. HFSR results
in significant morbidity, and dose modification including drug
discontinuation remain the only option for high grade HFSR.
References
- World Health
Organization [WHO]. Tuberculosis. Fact sheet 104. March 2012. Ed.,
2012.
- Azad A, Sadee W,
Schlesinger L. Innate
immune gene polymorphisms in tuberculosis. Infection and immunity.
2012; 80; 3343-3359. http://dx.doi.org/10.1128/IAI.00443-12

- Bellamy R, Beyers N,
McAdam K, Ruwende C,
Gie R, Samaai P, Bester D, Meyer M, Corrah T. Genetic susceptibility to
tuberculosis in Africans: a genome-wide scan. Proceedings of the
National Academy of Sciences. 2000; 97: 8005-8009. http://dx.doi.org/10.1073/pnas.140201897

- Sharma S, Rathored J,
Ghosh B, Sharma S.
Genetic polymorphisms in TNF genes and tuberculosis in North Indians.
BMC infectious diseases. 2010; 10 (1): 165. http://dx.doi.org/10.1186/1471-2334-10-165

- Moran A, Ma X, Reich R,
Graviss E. No
association between the 874T/A single nucleotide polymorphism in the
IFN-gene and susceptibility to TB. The International Journal of
Tuberculosis and Lung Disease. 2007; 11 (1): 113-115.

- Flynn J, Ernst J. Immune
responses in
tuberculosis. CurrOpin Immunol. 2000; 12 (4): 432-436. http://dx.doi.org/10.1016/S0952-7915(00)00116-3

- Farnia P, Masjedi M,
Mirsaeidi M, Mohammadi
F, Vincent V, Bahadori M, Velayati AA. Prevalence of Haarlem I and
Beijing types of Mycobacterium tuberculosis strains in Iranian and
Afghan MDR-TB patients. Journal of Infection. 2006; 53 (5): 331-336. http://dx.doi.org/10.1016/j.jinf.2005.12.020

- Selvaraj P, Sriram U,
Kurian S, Reetha A,
Narayanan P. Tumour necrosis factor alpha [-238 and -308] and beta gene
polymorphisms in pulmonary tuberculosis: haplotype analysis with HLA-A,
B and DR genes. Tuberculosis [Edinb]. 2001; 81 (5): 335-341. http://dx.doi.org/10.1054/tube.2001.0307

- Wang Q, Zhan P, Qiu L,
Qian Q, Yu L.
TNF-308 gene polymorphism and tuberculosis susceptibility: a
meta-analysis involving 18 studies. Molecular biology reports. 2012; 39
(4): 3393-3400. http://dx.doi.org/10.1007/s11033-011-1110-x

- Vejbaesya S, Chierakul
N, Luangtrakool P,
Sermduangprateep C. NRAMP1 and TNF-a polymorphisms and susceptibility
to tuberculosis in Thais. Respirology. 2007; 12 (2): 202-206. http://dx.doi.org/10.1111/j.1440-1843.2006.01037.x

- Lopez B, Aguilar D,
Orozco H, Burger M,
Espitia C, Ritacco V, Barrera L, Kremer K, Hernandez-pando R, Huygen K.
A marked difference in pathogenesis and immune response induced by
different Mycobacterium tuberculosis genotypes. Clinical &
Experimental Immunology. 2003; 133 (1): 30-37. http://dx.doi.org/10.1046/j.1365-2249.2003.02171.x

- Tanveer M, Hasan Z,
Kanji A, Hussain R,
Hasan R. Reduced TNF-a and IFN-? responses to Central Asian strain 1
and Beijing isolates of Mycobacterium tuberculosis in comparison with
H37Rv strain. Transactions of the Royal Society of Tropical Medicine
and Hygiene. 2009; 103 (6): 581-587. http://dx.doi.org/10.1016/j.trstmh.2009.03.014

- Rakotosamimanana N,
Raharimanga V,
Andriamandimby S, Soares J, Doherty T, Ratsitorahina M, Ramarokoto H,
Zumla A, Huggett J, Rook G. Variation in gamma interferon responses to
different infecting strains of Mycobacterium tuberculosis in acid-fast
bacillus smear-positive patients and household contacts in
Antananarivo, Madagascar. Clinical and Vaccine Immunology. 2010; 17
(10): 1094-1103. http://dx.doi.org/10.1128/CVI.00049-10

- Van Laarhoven A,
Mandemakers J,
Kleinnijenhuis J, Enaimi M, Lachmandas E, Joosten M, Ottenhoff T, Netea
M, Van Soolingen D, Van Crevel R. Low Induction of Proinflammatory
Cytokines Parallels Evolutionary Success of Modern Strains within the
Mycobacterium tuberculosis Beijing Genotype. Infection and immunity.
2013; 81 (10): 3750-3756. http://dx.doi.org/10.1128/IAI.00282-13

- Velayati A, Farnia P,
Mirsaeidi M, Masjedi
M .The most prevalent Mycobacterium tuberculosis superfamilies among
Iranian and Afghan TB cases. Scandinavian journal of infectious
diseases. 2006; 38 (6): 463-468. http://dx.doi.org/10.1080/00365540500504117

- Glynn J, Whiteley J,
Bifani P, Kremer K,
Van Soolingen D. Worldwide occurrence of Beijing/W strains of
Mycobacterium tuberculosis: a systematic review. Emerg Infect Dis.
2002; 8 (8): 843-849. http://dx.doi.org/10.3201/eid0805.020002

- Duchêne V, Ferdinand S,
Filliol I, Guégan
J, Rastogi N, Sola C. Phylogenetic reconstruction of Mycobacterium
tuberculosis within four settings of the Caribbean region: tree
comparative analyze and first appraisal on their phylogeography.
Infection, Genetics and Evolution. 2004; 4 (1): 5-14. http://dx.doi.org/10.1016/j.meegid.2003.09.001

- Caminero J, Pena M,
Campos-Herrero M,
Rodriguez J, Garcia I, Cabrera P, Lafoz C, Samper S, Takiff H, Afonso
O. Epidemiological evidence of the spread of a Mycobacterium
tuberculosis strain of the Beijing genotype on Gran Canaria Island.
American journal of respiratory and critical care medicine. 2001; 164
(7): 1165-1170. http://dx.doi.org/10.1164/ajrccm.164.7.2101031

- Petroff S. A new and
rapid method for the
isolation and cultivation of tubercle bacilli directly from the sputum
and feces. The Journal of experimental medicine. 1915; 21 (1): 38-42. http://dx.doi.org/10.1084/jem.21.1.38

- Kamerbeek J, Schouls L,
Kolk A, Van
Agterveld M, Van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H,
Shaw R, Goyal M. Simultaneous detection and strain differentiation of
Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin
Microbiol. 1997; 35 (4): 907-914.

- Rieder H, Chonde T,
Myking H, et al. The
public health service national tuberculosis reference laboratory and
the national laboratory network; minimum requirements, role and
operation in a low-income country. International Union against
Tuberculosis and Lung Disease [IUATLD], 1998.
- Merza M, Farnia P,
Anoosheh S, Varahram M,
Kazampour M, Pajand O, Saeif S, Mirsaeidi M, Masjedi MR, Velayati AA.
The NRAMPI, VDR and TNF-a gene polymorphisms in Iranian tuberculosis
patients: the study on host susceptibility. Brazilian Journal of
Infectious Diseases. 2009; 13 (4): 252-256. http://dx.doi.org/10.1590/S1413-86702009000400002

- Sambrook J, Russell DW.
Molecular cloning: A Laboratory Manual. CSHL Press 2001.
- Anoosheh S, Farnia P,
Kargar M. Association
between TNF-Alpha [-857] gene polymorphism and susceptibility to
tuberculosis. Iranian Red Crescent Medical Journal 2011; 13 (4); 243.

- Awomoyi A, Nejentsev S,
Richardson A, Hull
J, Koch O, Podinovskaia M, Todd J, McAdam K, Blackwell J, Kwiatkowski
D. No association between interferon-? receptor-1 gene polymorphism and
pulmonary tuberculosis in a Gambian population sample. Thorax. 2004; 59
(4); 291-294. http://dx.doi.org/10.1136/thx.2003.013029

- Bulat-Kardum L, Etokebe
G, Knezevic J,
Balen S, Matakovic-Mileusnic N, Zaputovic L, Pavelic J, Beg-Zec J,
Dembic Z. Interferon-? Receptor-1 Gene Promoter Polymorphisms [G-611A;
T-56C] and Susceptibility to Tuberculosis. Scandinavian journal of
immunology. 2006; 63 (2): 142-150. http://dx.doi.org/10.1111/j.1365-3083.2005.01694.x

- Van der Spuy G. Kremer
K, Ndabambi S,
Beyers N, Dunbar R, Marais B, Van Helden P, Warren R. Changing
Mycobacterium tuberculosis population highlights clade-specific
pathogenic characteristics. Tuberculosis. 2009; 89 (2): 120-125. http://dx.doi.org/10.1016/j.tube.2008.09.003

- Chakraborty P, Kulkarni
S, Rajan R, Sainis
K. Drug Resistant Clinical Isolates of Mycobacterium tuberculosis from
Different Genotypes Exhibit Differential Host Responses in THP-1 Cells.
PLoS One. 2013; 8 (5): e62966. http://dx.doi.org/10.1371/journal.pone.0062966

- Boeuf P, Vigan-Womas I,
Jublot D, Barale J,
Akanmori B, Mercereau-Puijalon O, Behr C. CyProQuant-PCR: a real time
RT-PCR technique for profiling human cytokines, based on external RNA
standards, readily automatable for clinical use. BMC immunology. 2005;
6 (1); 5. http://dx.doi.org/10.1186/1471-2172-6-5

- Amirzargar A, Rezaei N,
Jabbari H, Danesh
A, Khosravi F, Hajabdolbaghi M, Yalda A, Nikbin B. Cytokine single
nucleotide polymorphisms in Iranian patients with pulmonary
tuberculosis. European cytokine network. 2006; 17 (3): 84-89.

- Ates O, Musellim B,
Ongen G, Topal-Sarikaya
A. Interleukin-10 and tumor necrosis factor-a gene polymorphisms in
tuberculosis. Journal of clinical immunology. 2008; 28 (2): 232-236. http://dx.doi.org/10.1007/s10875-007-9155-2

- Perrey C, Turner S,
Pravica V, Howell W,
Hutchinson I. ARMS-PCR methodologies to determine IL-10, TNF-a, TNF-ß
and TGF-ß1 gene polymorphisms. Transplant immunology. 1999; 7 (2):
127-128. http://dx.doi.org/10.1016/S0966-3274(99)80030-6

- Oh J, Yang C, Noh Y,
Kweon Y, Jung S, Son
J, Kong S, Yoon J, Lee J, Kim H. Polymorphisms of interleukin-10 and
tumour necrosis factor-a genes are associated with newly diagnosed and
recurrent pulmonary tuberculosis. Respirology. 2007; 12 (4): 594-598. http://dx.doi.org/10.1111/j.1440-1843.2007.01108.x

- Krishnan N, Malaga W,
Constant P, Caws M,
Chau T, Salmons J, Lan N, Bang N, Daffé M, Young D. Mycobacterium
tuberculosis lineage influences innate immune response and virulence
and is associated with distinct cell envelope lipid profiles. PLoS One.
2011; 6 (9): e23870. http://dx.doi.org/10.1371/journal.pone.0023870

- Mirsaeidi S, Houshmand
M, Tabarsi P, Banoei
M, Zargari L, Amiri M, Mansouri S, Sanati M, Masjedi M. Lack of
association between interferon-gamma receptor-1 polymorphism and
pulmonary TB in Iranian population sample. Journal of Infection. 2006;
52 (5): 374-377. http://dx.doi.org/10.1016/j.jinf.2005.08.009

- Rook G, Steele J,
Ainsworth M, Champion B.
Activation of macrophages to inhibit proliferation of Mycobacterium
tuberculosis: comparison of the effects of recombinant gamma-interferon
on human monocytes and murine peritoneal macrophages. Immunology. 1986;
59 (3): 333, 198.

[TOP]








































