Birama Diarra1,2*, Albert Théophane Yonli1,2*, Pegdwendé Abel Sorgho1,2, Tegwindé Rebeca Compaore1,2, Abdoul Karim Ouattara1,2, Wendpagnangdé Arsène Zongo1,2, Issoufou Tao1,2, Lassina Traore1,2, Serge Théophile Soubeiga1,2, Florencia Wendkuuni Djigma1,2, Dorcas Obiri-Yeboah3, Bolni-Marius Nagalo1,2, Virginio Pietra1, Rokia Sanogo4 and Jacques Simpore1,2.
1 Biomolecular Research Center Pietro Annigoni (CERBA), BP 364 Ouagadougou 01, Burkina Faso.
2
Laboratory of Molecular Biology and Molecular Genetics (LABIOGENE)
UFR/SVT, University Ouaga I Prof Joseph KI-ZERBO, Burkina Faso; BP 7021
Ouagadougou 03.
3 Department of Microbiology and Immunology, School of Medical Sciences, University of Cape Coast, Ghana.
4 Faculty of Pharmacy, University of Sciences of Techniques and Technologies of Bamako (USTTB), Mali.
Corresponding
author: Birama Diarra, Laboratory of
Molecular Biology and Molecular Genetics (LABIOGENE) University Ouaga I
Prof Joseph KI-ZERBO, Burkina Faso. Telephone: +226 66643692/+223
79126274; E-mail:
diarra.birama679@gmail.com
Published: January 1, 2018
Received: September 7, 2017
Accepted: December 15, 2017
Mediterr J Hematol Infect Dis 2018, 10(1): e2018007 DOI
10.4084/MJHID.2018.007
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background: The
presence of HBV DNA in the liver (with detectable or undetectable HBV
DNA in the serum) of individuals tested HBsAg negative by currently
available assays is defined occult B Infection (OBI). It remains a
potential transmission threat and risk to HBV chronic infection. The
purpose of this study was to determine the OBI prevalence among HBsAg
negative subjects and to characterize associated genotypes.
Methods:
Blood samples of 219 HBsAg-negative subjects tested by ELISA were
collected. HBV DNA was investigated in all samples. Viral loads were
determined using quantitative real-time PCR. All samples were screened
for HBV markers (anti-HBc, anti-HBe, HBsAg). The Pre-S/S region of the
HBV genome was sequenced. The database was analyzed using the SPSS and
Epi info software. Phylogenetic analysis was performed using the
BioEdit and MEGA software.
Results:
Of the 219 samples, 20.1% were anti-HBc positive, 1.8% HBeAg and 22.8%
were anti-HBe positive. Fifty-six (56) (25.6%) of the samples had a
detectable HBV DNA and viral loads ranging from 4 IU/mL to 13.6 106
IU/mL. Sixteen of them (16/56) had a viral load < 200 IU/mL,
resulting in an OBI prevalence of 7.3% (16/219) in our study. The
remaining 40 subjects had viral loads ˃ 200 IU/mL, resulting in a
“false OBI” prevalence of 18.3% (40/219). HBV genotype E was
predominant followed by the quasi-sub-genotype A3. A single "false OBI"
strain had the characteristic mutation G145R. Other mutations were
observed and all located in the major hydrophilic region (MHR) of the S
gene.
Conclusion: The
study reported a prevalence of 7.3% of occult hepatitis B infection. It
confirms the predominance of genotype E and the existence of a subgroup
of quasi-sub-genotype A3 of HBV in Burkina Faso. It further provides
information on the presence of "false OBI." This study has found
mutations in the major hydrophilic region (MHR) of the pre-S/S gene of
HBV.
|
Introduction
Hepatitis
B virus (HBV) infection remains a major public health problem
worldwide. Approximately more than 360 million people are chronic
carriers of HBV, and more than 700,000 die each year from cirrhosis or
hepatocellular carcinoma.[1] HBV infection is highly endemic (prevalence ≥ 8% in the general population) in sub-Saharan Africa.[2]
Burkina Faso (BF) is a highly endemic country with prevalence F HBV between 10% - 15% in the general population.[3,4]
Some prevalences of 14.3%, 17%, and 12.9% has been reported among the
blood donors in Nouna, Ouagadougou and the National Blood Transfusion
Center of Burkina Faso respectively.[5,6] Moreover, prevalences of 9.3% and 9.8% has been reported among pregnant women in Burkina Faso.[7,8]
The
serological diagnosis of the hepatitis B virus (HBV) infection is
mainly based on tests for the detection of hepatitis B surface antigen
(HBsAg), and its absence is believed to exclude the occurrence of an
infection. The presence of HBV DNA in the liver (with detectable or
undetectable HBV DNA in the serum) of individuals tested HBsAg negative
by currently available assays is defined occult B Infection (OBI).[9] When detectable, the amount of HBV DNA in the serum is usually very low (< 2 00 IU/ml).[9]
The
detection of OBI has been reported among subjects with clinical
manifestations, such as chronic liver disease and hepatocellular
carcinoma.[10] Although most OBI carriers are asymptomatic, it has been detected in patients with chronic liver disease "cryptogenic"[11,12] and may be associated with progression towards liver fibrosis and cirrhosis development.[10]
Currently,
a maximum of ten genotypes (A-J) and several sub-genotypes of HBV with
a distinct geographical distribution have been characterized.[13,14]
Several studies have shown that the clinical picture, treatment
response, long-term prognosis and seroconversion profile are influenced
by HBV genotypes.[15,16]
In Burkina Faso, very
few studies have focused on occult HBV infection and associated
genotypes. However, a recent study reported a prevalence of 32.8 %
(25/76) of OBI among blood donors of Ouagadougou.[17]
Thus, this study aimed to determine the prevalence of OBI among HBsAg
negative subjects and characterize the associated genotypes.
Methods
Ethical consideration.
Approval for the study was obtained from the National Health Ethics
Committee of Burkina Faso (reference number 2015-6-080 of June, 10th
2015). Informed consent was obtained from all participants before blood
collection in accordance with the Helsinki Declarations
Study population.
The study was conducted between October 2014 and January 2017 in
Ouagadougou, at the Pietro Annigoni Biomolecular Research Center (CERBA
/ LABIOGENE) of Burkina Faso. The study population consisted of 219
HBsAg-negative subjects and non-vaccinated against hepatitis B,
regardless of age or social category. Participants were recruited
following an awareness campaign on hepatitis and sociodemographic
characteristics registered.
Sample collection, HBsAg serology, and HBV markers.
The sampling was preceded by an awareness campaign on the transmission
modes, risk groups, the symptoms, complications, the importance of
screening and the means of prevention against hepatitis B. Blood
samples collected from 219 subjects were centrifuged, and plasmas were
stored at -20°C until use. HBsAg was tested using the ELISA method on
the Cobas e 411 Analyzer (Roche Diagnostics GmbH Mannheim Germany) with
a lower detection limit of 0.05 UI/mL. HBV markers (anti-HBc, anti-HBc,
HBeAg) were determined among all participants using the same device.
DNA extraction.
Viral DNA was extracted from 200μL of serum samples using QIAamp DNA
Blood Mini kit (Qiagen GmbH, Hilden, Germany) following the
manufacturer’s instructions and was stored at - 20°C until use.
Quantification of HBV DNA.
The quantification of the HBV-DNA was performed using the 7 500
Real-Time PCR System (Applied Biosystems, USA). The target gene was a
highly conserved region of surface gene provides for the accurate
detection of genotypes A-H. The HBV-plasmid DNA was used to generate a
standard curve following a serial 10-fold dilution. Our quantitative
HBV-specific PCR assays were routinely standardized using the WHO
standard (NIBSC code: 97/750).
Amplification and sequencing of HBV DNA.
The pre-S/S region of the HBV genome of 21 samples was amplified using
nested PCR and directly sequenced according to the method of Chen et
al., 2007.[18] The detection limit of the HBV DNA was
20 IU/mL. Molecular cloning and sequencing were performed only when
pre-S deletions were found by direct sequencing. The HBV pre-S/S gene
PCR products were cloned into the TOPO®TA cloning kit (Invitrogen Ltd,
Paisley, UK) according to the manufacturer’s instructions. Plasmid DNA
from clones was purified with the GFX PCR purification kit (Healthcare,
Buckinghamshire, UK) and sequenced. Sequencing was performed using the
BigDye Terminator cycle sequencing kit (Applied Biosystems, CA, USA)
and analyzed on the ABI PRISM Genetic Analyzer 3130XL (Applied
Biosystems, CA, USA) according to manufacturer’s instructions.
Statistical and phylogenetic analysis.
The data were analyzed using the SPSS 21.0 and Epi Info version 7.0
software. The chi-square test was used for the comparisons, and the
difference was considered statistically significant for p ≤ 0.05.
Sequencing results were analyzed using BioEdit 7.2.6 software. Multiple
sequence alignment was performed with Clustal W software on HBV
sequences of genotypes A–H available in GenBank (http://www.ncbi.nlm.nih.gov/genbank/index.htm).
Phylogenetic analysis was performed using the Kimura two-parameter
model and tree were constructed with neighbor-joining and maximum
likelihood methods using the MEGA software version 5.1. Results
Demographic and serologic characteristics of the study population.
A total of 219 individuals, aged between 14 and 77 years (mean age of
38.4 ± 13.5 years), including 102 (46.6%) women and 117 (53.4%) men
participated in this study. The most represented age group was 31 to 40
years, with 52.1% (114/219). Of the 219 HBsAg-negative individuals, 44
(20.1%) were anti-HBc positive, 3 (1.8%) HBeAg positive and 50 (22.8%)
anti-HBe positive (Table 1). However, 56 (25.6%) of the samples had detectable HBV DNA by real-time PCR using HBV-specific primer pairs.
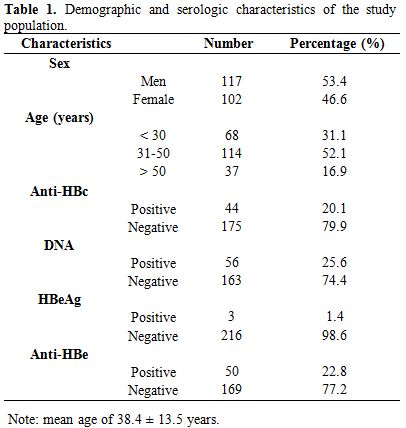 |
Table 1. Demographic and serologic characteristics of the study population. |
Characteristics of samples with viral DNA of HBV (n = 56) according to their viral loads. Of the 56 samples with HBV DNA, 32 (57.1%) were women and 24 (42.9%) men (Table 2).
HBV DNA was quantified in the 56 samples by real-time PCR, of which
78.5% (44/56) were anti-HBc-positive. Their viral loads ranged from 4
IU/mL to 13.6 106 UI/mL. An occult
hepatitis B virus infection (OBI) prevalence of 7.3% (16/219) was
observed in this study. The majority of OBI carriers were anti-HBc
positive (14/16) and mainly constituted of men (9/16) in the age group
31-50 (Table 2). In general,
the prevalence of HBV markers was 12.5%, 87.5% and 12.5% for anti-HBs,
anti-HBc, and anti-HBe respectively. These prevalences were mostly
higher in samples with a viral load ˃ 200 IU/mL (Table 2).
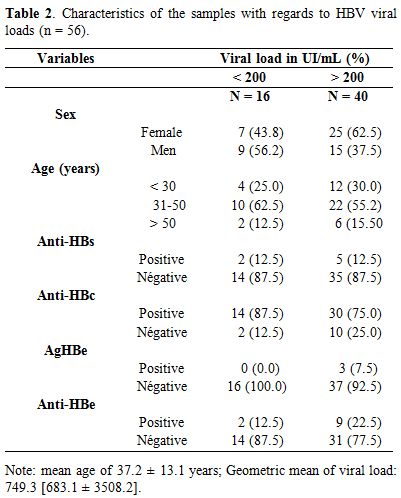 |
Table 2. Characteristics of the samples with regards to HBV viral loads (n = 56). |
Sequencing and determination of HBV genotypes.
The 21 pre-S/S HBV sequences of the present study were analyzed
together with 208 sequences of genotype E and A3 African strains
available in the GenBank database. Both neighbor-joining and maximum
likelihood phylogenetic reconstructions showed that our sequences and
the previously characterized African HBV genotypes E and A3 sequences
were dispersed within clade E irrespective of their geographical
origins (Figure 1). Also, the
HBV genotypes E, and A3 sequences of the present study were clustered
precisely within the same clade E and A3 respectively among the
Burkinabe sequences previously deposited in GenBank (Figure 1).
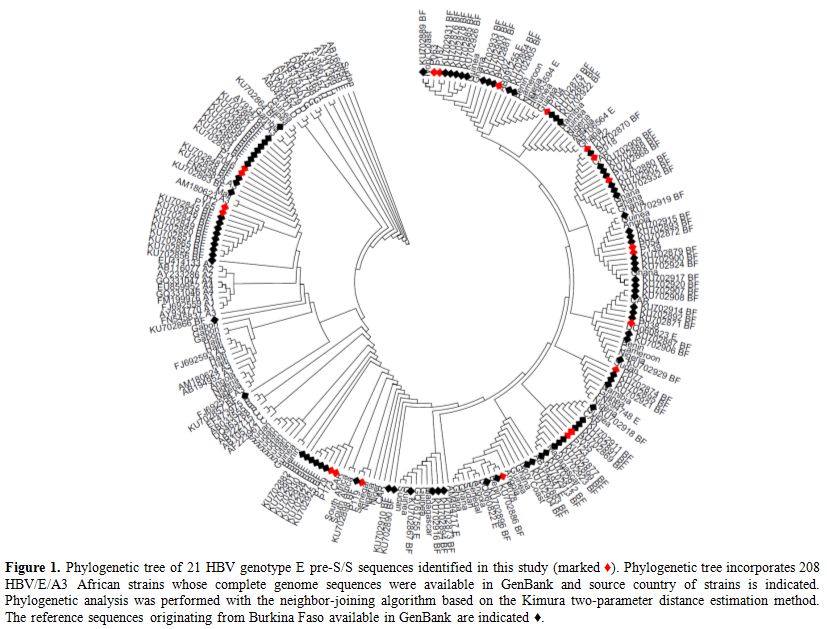 |
Figure 1. Phylogenetic tree of 21 HBV
genotype E pre-S/S sequences identified in this study (marked ♦).
Phylogenetic tree incorporates 208 HBV/E/A3 African strains whose
complete genome sequences were available in GenBank and source country
of strains is indicated. Phylogenetic analysis was performed with the
neighbor-joining algorithm based on the Kimura two-parameter distance
estimation method. The reference sequences originating from Burkina
Faso available in GenBank are indicated ♦. |
The
HBV genome pre-S/S region of 16 OBI and 5 “false OBI” (21) samples were
sequenced. All sequences were considered for phylogenetic analysis and
genotyping (Figure 2). Four
sequences were clustered with HBV genotype A, and 17 sequences with
genotype E supported by 75% and 67% bootstrapping for 1,000 replicates,
respectively. The HBV genotype E pre-S/S sequences (n = 17) were
analyzed together with 67 sequences of Burkinabe strains and 44
references sequences including 9 of genotype E, all available in
GenBank. Both neighbor-joining and maximum likelihood phylogenetic
reconstructions showed that the 17 sequences were clustered within the
same clade E of the Burkinabe HBV genotype E sequences previously
characterized (Figure 2).
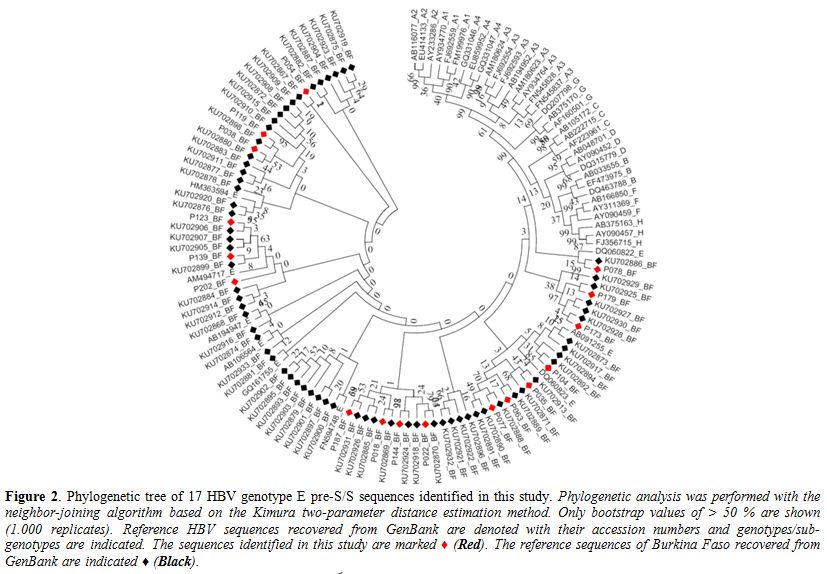 |
Figure 2. Phylogenetic tree of 17 HBV genotype E pre-S/S sequences identified in this study. Phylogenetic
analysis was performed with the neighbor-joining algorithm based on the
Kimura two-parameter distance estimation method. Only bootstrap values
of > 50 % are shown (1.000 replicates). Reference HBV sequences
recovered from GenBank are denoted with their accession numbers and
genotypes/sub-genotypes are indicated. The sequences identified in this
study are marked ♦ (Red). The reference sequences of Burkina Faso
recovered from GenBank are indicated ♦ (Black). |
Also,
the HBV genotype A pre-S/S sequence (n=4) were analyzed together with
22 A3 sub-genotype sequences of Burkinabe strains and 44 references
sequences including 8 of A3 sub-genotype, all available in GenBank.
Phylogenetic analysis also showed that the 4 sequences were HBV subtype
A3 and clustered in same clade A3 (Figure 3).
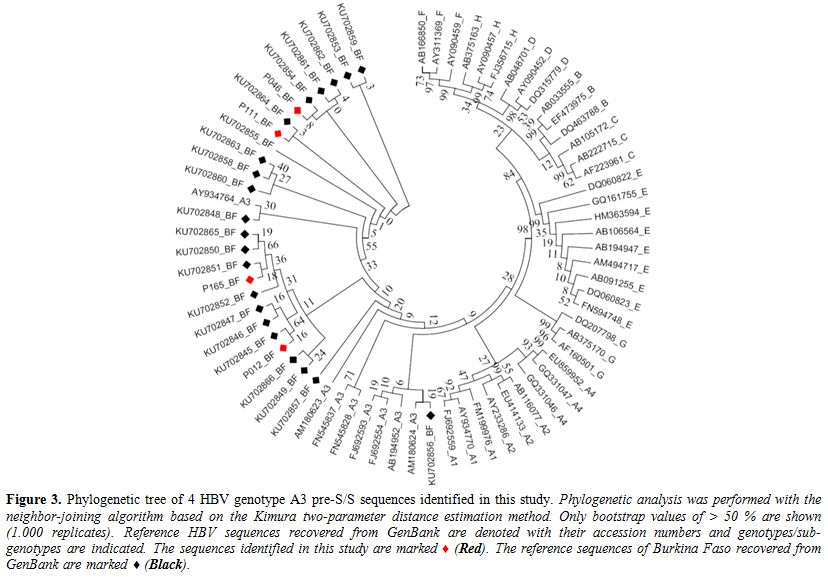 |
Figure 3. Phylogenetic tree of 4 HBV genotype A3 pre-S/S sequences identified in this study. Phylogenetic
analysis was performed with the neighbor-joining algorithm based on the
Kimura two-parameter distance estimation method. Only bootstrap values
of > 50 % are shown (1.000 replicates). Reference HBV sequences
recovered from GenBank are denoted with their accession numbers and
genotypes/sub-genotypes are indicated. The sequences identified in this
study are marked ♦ (Red). The reference sequences of Burkina Faso
recovered from GenBank are marked ♦ (Black). |
Mutations in the S gene according to genotypes and cases of hepatitis B virus infection.
Of the 21 pre-S/S regions sequenced, 16 (76.2 %) were OBI cases and 5
(23.8 %) "false OBI" cases. The A3 genotype strains showed no specific
mutations. A single strain of “false OBI “carried the G145R mutation (Table 3). All other amino acid substitutions were observed in both cases (Table 3). In general, all observed mutations are located in the most hydrophilic region (MHR) of the S gene (Table 3).
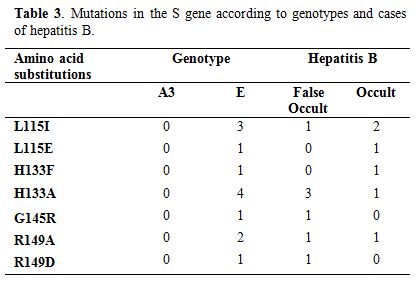 |
Table 3. Mutations in the S gene according to genotypes and cases of hepatitis B. |
Discussion
In
this study, the anti-HBc prevalence was 20.1% (44/219) among
HBsAg-negative subjects. This prevalence is lower than 44.0% reported
in HBsAg-negative blood donors in Burkina Faso.[17] However, it is higher than 7.8% and 16.6% reported in HBsAg-negative blood donors in Egypt.[19,20]
These differences could be explained by the size and type of study
population but also by endemicity for HBV. It should also be mentioned
that voluntary participation in a screening program includes
self-selection bias.
Until now, most studies of occult hepatitis B
virus infection were conducted among blood donors, poly-transfused
patients or patients with proven or co-infected with liver disease.
Data on the prevalence of OBI is limited in sub-Saharan Africa, in
particular among alleged healthy individuals. The prevalence of occult
HBV infection was 7.3% in our study. The latter is lower than that
reported among HIV-positive patients from Ivory Coast in 2010 and from
Sudan in 2014, and among blood donors from Burkina Faso in 2016; 10%,
15%, and 32.8% respectively.[17,21,22] Nevertheless, our prevalence was similar to that of 6.25% reported among Egyptian blood donors in 2010.[19] However, it was higher than 0.5% reported among regular blood donors in Southeast Nigeria.[23]
These variations could be explained by the difference of population
studied, the sensitivity of the diagnostic tests used and the
prevalence of HBV. Indeed, several studies have shown that OBI is
significantly associated with the endemicity of HBV infection but not
restricted to countries which are highly endemic to the virus.[24,25]
Thus, assays that use polyclonal antibodies show higher sensitivity and
specificity for the detection of various types of HBsAg mutants than
those using monoclonal antibodies.[26,27] It is also
worth noting that the nature of the specimen tested (i.e., a blood
sample or liver tissue), the amount of specimen, as well as
contamination risks, can also affect the detection of OBI.[28]
A
low level of HBV viral load (< 200 IU/mL) was observed among OBI
cases in this study. Indeed, several studies have shown that almost all
OBI cases are infected with replication-competent HBV, revealing a
strong suppression of replication activity and gene expression,
therefore resulting in a reduced viral load.[9,29,30]
Other studies have also shown that a limited number of OBI cases are
due to infection with HBV mutants with defective replication activity
or S protein synthesis.[31,32] It was also reported that HBV DNA could integrate into the OBI host genome.[29,33]
In this study, more than two-thirds of subjects with HBV DNA (40/56) had a viral load ˃ 200 IU/mL (200 to 13.6 106
IU/mL). This could be attributed to escape mutations that can lead to a
change in the immunologic epitope thus inhibiting HBsAg secretion.[34]
This hypothesis is based on a small number of sequenced HBV-DNA and
needs further confirmation. A study reported a viral load between
undetectable and 3,670 IU/mL in "OBI" cases among blood donors in
Southeast Asia.[35] In 2008, the statements from the
Taormina expert meeting on occult hepatitis B virus infection had
clarified the definition of OBI in establishing a threshold value of
serum HBV DNA < 200 IU/mL.[9] Furthermore, it also
clarified the confusion between a cleared infection of HBV and a "false
OBI". Thus, cases with serum HBV DNA levels comparable to those usually
detected in the different phases of serologically evident (overt) HBV
infection have to be considered as "false OBI" and are usually due to
infection by HBV variants.[9] These become in fact
chronic hepatitis B cases. We believe that not taking these definitions
into account may contribute to an overestimation of the prevalence of
OBI.
HBV Genotype E was most prevalent in OBI cases in this study.
The HBV genotype E sequences of this study were similar to those
previously characterized in Burkina Faso.[13] These results confirm the endemicity and low genetic diversity of HBV genotype E in West Africa.[36] In addition, HBV sub-genotype A3, previously reported in Burkina Faso,[13]
was also observed in this study. This result confirms those of previous
studies which have shown that HBV sub-genotype A3 and recombination
between HBV genotypes A and E are frequently observed in West Africa.[13,37,38]
In
this study, the L115I/A; H133F/A, and R149A/D mutations were found in
OBI cases. However, the results of previous studies have reported that
the Pre-S/S gene has a relatively high mutation rate.[28]
These point mutations that occur in the Pre-S/S gene may affect
antigenicity, immunogenicity, secretion, and/or expression of HBsAg,
leading to detection failure of HBsAg.[26,39] They may also reduce or even abolish the replication and/or secretion of the virion, exerting an adverse effect on HBsAg.[40,41]
It was also reported that amino acid (aa) substitutions of HBsAg are
frequently clustered in the “α” determinant, which is located at the
position aa124-147 of the S protein.[28] This
determinant "α" is a relatively conserved region within the major
hydrophilic region (MHR) between aa100 to aa169, which serves as the
most important antigenic determinant in all HBV strains and is
essential to the detection of HBsAg and development of HBV vaccines.[42,43] Amino acids within the region aa120 to 123 were shown to be crucial for the antigenicity of HBsAg.[44]
Therefore, single or multiple point mutations occurring within or
adjacent to the "α" determinant may change the antigenicity and
conformation of HBsAg, failing to detect HBsAg.[28] The results of a recent study suggest that HBsAg variants may not play a major role in OBI pathogenesis.[45]
All mutations characterized in this study were located in the major
hydrophilic region (MHR) of the S gene and could explain the nature of
occult HBV infection in our study. In addition, the same mutations were
observed in the "false OBI "cases.
The presence of same
mutations in addition to that of G145R in "false OBI" cases of this
study confirms the conclusion of the statements from the Taormina
expert meeting on occult hepatitis B virus infection.[9]
Indeed, in "false OBI" the viral load is similar to that of chronic
hepatitis B. In addition, the role of the G145R mutation has been
clearly established by several studies in vaccine escape.[41]
This study not found more than one type of escape mutation in the same
sample. Further studies are needed to confirm the mutations found in
this study.
Conclusions
In
conclusion, this study reported a prevalence of occult HBV infection of
7.3% among HBsAg seronegative patients in Burkina Faso. It confirms the
predominance and low HBV genotype E genetic diversity in West Africa.
It also established the existence a clade HBV sub-genotype A3 in
Burkina Faso. Our study also provided information on the presence a
"false OBI". The mutations observed in the MHR region of pre-S/S gene
may explain the occult nature of HBV infection in our study.
Acknowledgements
The
authors wish to thank the Laboratory of Molecular Biology and Genetics
(LABIOGENE) UFR/SVT, University Ouaga I Prof Joseph KI-ZERBO, Burkina
Faso and the Biomolecular Research Center Pietro Annigoni of
Ouagadougou (CERBA).
References
- Organization. WH. Global hepatitis report, 2017. www.who.int/hepatitis/publications/global-hepatitis-report2017/en, Accessed 24 April 2017.
- Birama.
D, Karim. OA, Wendkuuni. DF, Rebeca. CT, OBIRI-YEBOAH. D, Lassina. T,
Théophile. SS, Prosper. B, Justine. Y, Virginio. P, Paul. O, Alain. B,
. SR. Jacques. S. World Hepatitis Day 2016 in Burkina Faso: Awareness,
Screening, Identification of Hepatitis B Markers, HBV/HCV co-infection
and vaccination. Hepat Mon. 2017, 17(6):e13789. doi:
10.5812/hepatmon.13789.

- Burnett
RJ, Francois G, Kew MC, Leroux-Roels G, Meheus A, Hoosen AA. Mphahlele
MJ. Hepatitis B virus and human immunodeficiency virus co-infection in
sub-Saharan Africa: a call for further investigation. Liver Int. 2005,
25(2):201-213. https://doi.org/10.1111/j.1478-3231.2005.01054.x PMid:15780040

- Tao
I, Compaore TR, Diarra B, Djigma F, Zohoncon TM, Assih M, Ouermi D,
Pietra V, Karou SD. Simpore J. Seroepidemiology of hepatitis B and C
viruses in the general population of burkina faso. Hepat Res Treat.
2014, 2014:781843
 .
.
- Collenberg
E, Ouedraogo T, Ganame J, Fickenscher H, Kynast-Wolf G, Becher H,
Kouyate B, Krausslich HG, Sangare L. Tebit DM. Seroprevalence of six
different viruses among pregnant women and blood donors in rural and
urban Burkina Faso: A comparative analysis. J Med Virol. 2006,
78(5):683-692. https://doi.org/10.1002/jmv.20593 PMid:16555290

- Tao
I, Bisseye C, Nagalo BM, Sanou M, Kiba A, Surat G, Compaore TR, Traore
L, Nikiema JB, Pietra V, Zongo JD. Simpore J. Screening of Hepatitis G
and Epstein-Barr Viruses Among Voluntary non Remunerated Blood Donors
(VNRBD) in Burkina Faso, West Africa. Mediterr J Hematol Infect Dis.
2013, 5(1):e2013053. https://doi.org/10.4084/mjhid.2013.053 PMid:24106603 PMCid:PMC3787664

- Simpore
J, Granato M, Santarelli R, Nsme RA, Coluzzi M, Pietra V, Pignatelli S,
Bere A, Faggioni A. Angeloni A. Prevalence of infection by HHV-8, HIV,
HCV and HBV among pregnant women in Burkina Faso. J Clin Virol. 2004,
31(1):78-80. https://doi.org/10.1016/j.jcv.2004.06.001 PMid:15288619

- Simpore
J, Savadogo A, Ilboudo D, Nadambega MC, Esposito M, Yara J, Pignatelli
S, Pietra V. Musumeci S. Toxoplasma gondii, HCV, and HBV seroprevalence
and co-infection among HIV-positive and -negative pregnant women in
Burkina Faso. J Med Virol. 2006, 78(6):730-733. https://doi.org/10.1002/jmv.20615 PMid:16628587

- Raimondo
G, Allain JP, Brunetto MR, Buendia MA, Chen DS, Colombo M, Craxi A,
Donato F, Ferrari C, Gaeta GB, Gerlich WH, Levrero M, Locarnini S,
Michalak T, Mondelli MU, Pawlotsky JM, Pollicino T, Prati D, Puoti M,
Samuel D, Shouval D, Smedile A, Squadrito G, Trepo C, Villa E, Will H,
Zanetti AR. Zoulim F. Statements from the Taormina expert meeting on
occult hepatitis B virus infection. J Hepatol. 2008, 49(4):652-657. https://doi.org/10.1016/j.jhep.2008.07.014 PMid:18715666

- Said ZN. An overview of occult hepatitis B virus infection. World J Gastroenterol. 2011, 17(15):1927-1938. https://doi.org/10.3748/wjg.v17.i15.1927 PMid:21528070 PMCid:PMC3082745

- Castillo
I, Rodriguez-Inigo E, Lopez-Alcorocho JM, Bartolome J, Pardo M. Carreno
V. Comparative study on the clinical and virological characteristics
among patients with single occult hepatitis B virus (HBV), single
occult hepatitis C virus (HCV) and occult HBV and HCV dual infection. J
Med Virol. 2007, 79(3):236-241. https://doi.org/10.1002/jmv.20784 PMid:17245725

- Chemin
I, Zoulim F, Merle P, Arkhis A, Chevallier M, Kay A, Cova L, Chevallier
P, Mandrand B. Trepo C. High incidence of hepatitis B infections among
chronic hepatitis cases of unknown aetiology. J Hepatol. 2001,
34(3):447-454. https://doi.org/10.1016/S0168-8278(00)00100-8

- Candotti
D, Diarra B, Bisseye C, Tao I, Pham Quang K, Sanou M, Laperche S,
Sanogo R, Allain JP. Simpore J. Molecular characterization of hepatitis
B virus in blood donors from Burkina Faso: Prevalence of
quasi-subgenotype A3, genotype E, and mixed infections. J Med Virol.
2016, 88(12):2145-2156. https://doi.org/10.1002/jmv.24589 PMid:27253483

- Tatematsu
K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T,
Wakuta M, Miyakawa Y. Mizokami M. A genetic variant of hepatitis B
virus divergent from known human and ape genotypes isolated from a
Japanese patient and provisionally assigned to new genotype J. J Virol.
2009, 83(20):10538-10547. https://doi.org/10.1128/JVI.00462-09 PMid:19640977 PMCid:PMC2753143

- Ghosh
S, Banerjee P, Deny P, Mondal RK, Nandi M, Roychoudhury A, Das K,
Banerjee S, Santra A, Zoulim F, Chowdhury A. Datta S. New HBV
subgenotype D9, a novel D/C recombinant, identified in patients with
chronic HBeAg-negative infection in Eastern India. J Viral Hepat. 2013,
20(3):209-218. https://doi.org/10.1111/j.1365-2893.2012.01655.x PMid:23383660

- Lu
JJ, Chen EQ, Yang JH, Zhou TY, Liu L. Tang H. A mutation in the
interferon regulatory element of HBV may influence the response of
interferon treatment in chronic hepatitis B patients. Virol J. 2012,
9:10. https://doi.org/10.1186/1743-422X-9-10 PMid:22233973 PMCid:PMC3287143

- Somda
KS, Sermé AK, Coulibaly A, Cissé K, Sawadogo A, Sombié AR. Bougouma A.
Hepatitis B Surface Antigen Should Not Be the Only Sought Marker to
Distinguish Blood Donors towards Hepatitis B Virus Infection in High
Prevalence Area. Open Journal of Gastroenterology. 2016, 6:200-210. https://doi.org/10.4236/ojgas.2016.611039

- Chen
CH, Hung CH, Lee CM, Hu TH, Wang JH, Wang JC, Lu SN. Changchien CS.
Pre-S deletion and complex mutations of hepatitis B virus related to
advanced liver disease in HBeAg-negative patients. Gastroenterology.
2007, 133(5):1466-1474. https://doi.org/10.1053/j.gastro.2007.09.002 PMid:17915220

- Antar
W, El-Shokry MH, Abd El Hamid WA. Helmy MF. Significance of detecting
anti-HBc among Egyptian male blood donors negative for HBsAg. Transfus
Med. 2010, 20(6):409-413. https://doi.org/10.1111/j.1365-3148.2010.01021.x PMid:20573069

- Said
ZN, Sayed MH, Salama, II, Aboel-Magd EK, Mahmoud MH, Setouhy ME,
Mouftah F, Azzab MB, Goubran H, Bassili A. Esmat GE. Occult hepatitis B
virus infection among Egyptian blood donors. World J Hepatol. 2013,
5(2):64-73. https://doi.org/10.4254/wjh.v5.i2.64 PMid:23646231 PMCid:PMC3642725

- Mudawi
H, Hussein W, Mukhtar M, Yousif M, Nemeri O, Glebe D. Kramvis A. Overt
and occult hepatitis B virus infection in adult Sudanese HIV patients.
Int J Infect Dis. 2014, 29:65-70. https://doi.org/10.1016/j.ijid.2014.07.004 PMid:25449238

- N'Dri-Yoman
T, Anglaret X, Messou E, Attia A, Polneau S, Toni T, Chenal H, Seyler
C, Gabillard D, Wakasugi N, Eholie S. Danel C. Occult HBV infection in
untreated HIV-infected adults in Cote d'Ivoire. Antivir Ther. 2010,
15(7):1029-1034. https://doi.org/10.3851/IMP1641 PMid:21041918

- Nna
E, Mbamalu C. Ekejindu I. Occult hepatitis B viral infection among
blood donors in South-Eastern Nigeria. Pathog Glob Health. 2014,
108(5):223-228. https://doi.org/10.1179/2047773214Y.0000000144 PMid:24995918 PMCid:PMC4153823

- Gutierrez-Garcia
ML, Fernandez-Rodriguez CM, Lledo-Navarro JL. Buhigas-Garcia I.
Prevalence of occult hepatitis B virus infection. World J
Gastroenterol. 2011, 17(12):1538-1542. https://doi.org/10.3748/wjg.v17.i12.1538 PMid:21472117 PMCid:PMC3070122

- Yuen
MF, Lee CK, Wong DK, Fung J, Hung I, Hsu A, But DY, Cheung TK, Chan P,
Yuen JC, Fung FK, Seto WK, Lin CK. Lai CL. Prevalence of occult
hepatitis B infection in a highly endemic area for chronic hepatitis B:
a study of a large blood donor population. Gut. 2010, 59(10):1389-1393.
https://doi.org/10.1136/gut.2010.209148 PMid:20675695

- Ireland
JH, O'Donnell B, Basuni AA, Kean JD, Wallace LA, Lau GK. Carman WF.
Reactivity of 13 in vitro expressed hepatitis B surface antigen
variants in 7 commercial diagnostic assays. Hepatology. 2000,
31(5):1176-1182. https://doi.org/10.1053/he.2000.6407 PMid:10796895

- Weber
B. Diagnostic impact of the genetic variability of the hepatitis B
virus surface antigen gene. J Med Virol. 2006, 78 Suppl 1:S59-65. https://doi.org/10.1002/jmv.20610 PMid:16622880

- Zhu
HL, Li X, Li J. Zhang ZH. Genetic variation of occult hepatitis B virus
infection. World J Gastroenterol. 2016, 22(13):3531-3546. https://doi.org/10.3748/wjg.v22.i13.3531 PMid:27053845 PMCid:PMC4814639

- Brechot
C, Thiers V, Kremsdorf D, Nalpas B, Pol S. Paterlini-Brechot P.
Persistent hepatitis B virus infection in subjects without hepatitis B
surface antigen: clinically significant or purely "occult"? Hepatology.
2001, 34(1):194-203. https://doi.org/10.1053/jhep.2001.25172 PMid:11431751

- Vivekanandan
P, Kannangai R, Ray SC, Thomas DL. Torbenson M. Comprehensive genetic
and epigenetic analysis of occult hepatitis B from liver tissue
samples. Clin Infect Dis. 2008, 46(8):1227-1236. https://doi.org/10.1086/529437 PMid:18444860 PMCid:PMC3140175

- Blum
HE, Galun E, Liang TJ, von Weizsacker F. Wands JR. Naturally occurring
missense mutation in the polymerase gene terminating hepatitis B virus
replication. J Virol. 1991, 65(4):1836-1842. PMid:2002544
PMCid:PMC239993

- Chaudhuri
V, Tayal R, Nayak B, Acharya SK. Panda SK. Occult hepatitis B virus
infection in chronic liver disease: full-length genome and analysis of
mutant surface promoter. Gastroenterology. 2004, 127(5):1356-1371. https://doi.org/10.1053/j.gastro.2004.08.003 PMid:15521005

- Brechot
C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma:
old and new paradigms. Gastroenterology. 2004, 127(5 Suppl 1):S56-61. https://doi.org/10.1053/j.gastro.2004.09.016 PMid:15508104

- Bremer
CM, Saniewski M, Wend UC, Torres P, Lelie N, Gerlich WH. Glebe D.
Transient occult hepatitis B virus infection in a blood donor with high
viremia. Transfusion. 2009, 49(8):1621-1629. https://doi.org/10.1111/j.1537-2995.2009.02188.x PMid:19413737

- Candotti
D, Lin CK, Belkhiri D, Sakuldamrongpanich T, Biswas S, Lin S, Teo D,
Ayob Y. Allain JP. Occult hepatitis B infection in blood donors from
South East Asia: molecular characterisation and potential mechanisms of
occurrence. Gut. 2012, 61(12):1744-1753. https://doi.org/10.1136/gutjnl-2011-301281 PMid:22267593

- Mulders
MN, Venard V, Njayou M, Edorh AP, Bola Oyefolu AO, Kehinde MO, Muyembe
Tamfum JJ, Nebie YK, Maiga I, Ammerlaan W, Fack F, Omilabu SA, Le Faou
A. Muller CP. Low genetic diversity despite hyperendemicity of
hepatitis B virus genotype E throughout West Africa. J Infect Dis.
2004, 190(2):400-408. https://doi.org/10.1086/421502 PMid:15216479

- Kurbanov
F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N,
Ngansop C, Kaptue L, Miura T, Ido E, Hayami M, Ichimura H. Mizokami M.
A new subtype (subgenotype) Ac (A3) of hepatitis B virus and
recombination between genotypes A and E in Cameroon. J Gen Virol. 2005,
86(Pt 7):2047-2056. https://doi.org/10.1099/vir.0.80922-0 PMid:15958684

- Makuwa
M, Souquiere S, Telfer P, Apetrei C, Vray M, Bedjabaga I,
Mouinga-Ondeme A, Onanga R, Marx PA, Kazanji M, Roques P. Simon F.
Identification of hepatitis B virus subgenotype A3 in rural Gabon. J
Med Virol. 2006, 78(9):1175-1184. https://doi.org/10.1002/jmv.20678 PMid:16847965

- Hsu
CW. Yeh CT. Emergence of hepatitis B virus S gene mutants in patients
experiencing hepatitis B surface antigen seroconversion after
peginterferon therapy. Hepatology. 2011, 54(1):101-108. https://doi.org/10.1002/hep.24363 PMid:21503942

- Huang
CH, Yuan Q, Chen PJ, Zhang YL, Chen CR, Zheng QB, Yeh SH, Yu H, Xue Y,
Chen YX, Liu PG, Ge SX, Zhang J. Xia NS. Influence of mutations in
hepatitis B virus surface protein on viral antigenicity and phenotype
in occult HBV strains from blood donors. J Hepatol. 2012,
57(4):720-729. https://doi.org/10.1016/j.jhep.2012.05.009 PMid:22634131

- Kalinina
T, Iwanski A, Will H. Sterneck M. Deficiency in virion secretion and
decreased stability of the hepatitis B virus immune escape mutant
G145R. Hepatology. 2003, 38(5):1274-1281. https://doi.org/10.1053/jhep.2003.50484 PMid:14578867

- Norder
H, Courouce AM. Magnius LO. Molecular basis of hepatitis B virus
serotype variations within the four major subtypes. J Gen Virol. 1992,
73 (Pt 12):3141-3145. https://doi.org/10.1099/0022-1317-73-12-3141 PMid:1469353

- Seeger C. Mason WS. Hepatitis B virus biology. Microbiol Mol Biol Rev. 2000, 64(1):51-68. https://doi.org/10.1128/MMBR.64.1.51-68.2000 PMid:10704474

- Tian
Y, Xu Y, Zhang Z, Meng Z, Qin L, Lu M. Yang D. The amino Acid residues
at positions 120 to 123 are crucial for the antigenicity of hepatitis B
surface antigen. J Clin Microbiol. 2007, 45(9):2971-2978. https://doi.org/10.1128/JCM.00508-07 PMid:17609325 PMCid:PMC2045265

- Zhang
Z, Zhang L, Dai Y, Zhang Y, Li J. Li X. Occult hepatitis B virus
infection: influence of S protein variants. Virol J. 2016, 13:10. https://doi.org/10.1186/s12985-016-0464-zPMid:26786229 PMCid:PMC4717550

[TOP]







 .
. 







































