Mahtab Hadadi1, Hamid Heidari1, Hadi Sedigh Ebrahim-Saraie1 and Mohammad Motamedifar1,2.
1 Department of Bacteriology and Virology, School of Medicine, Shiraz University of Medical Sciences, Shiraz, Iran.
2 Shiraz HIV/AIDS Research Center, Institute of Health, Shiraz University of Medical Sciences, Shiraz, Iran.
Correspondence to: Department of Bacteriology & Virology, School of
Medicine, Shiraz University of Medical Sciences, Zand St, Imam Hossein
Sq, Shiraz, Iran. Tel/Fax: +98 713 2304356; E- mail:
motamedm@yahoo.com;
motamedm@sums.ac.ir
Published: September 1, 2018
Received: April 12, 2018
Accepted: June 18, 2018
Mediterr J Hematol Infect Dis 2018, 10(1): e2018053 DOI
10.4084/MJHID.2018.053
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background. Staphylococcus aureus
is a common cause of nosocomial infections leading to a broad spectrum
of diseases. Increasing antibiotic resistance among S.aureus strains, particularly methicillin-resistant S.aureus
(MRSA), is a serious concern. In addition, the emergence of antiseptics
resistance in MRSA helps the organism to persist and spread in
healthcare environments easily. The aim of this study was to determine
the molecular characteristics of vancomycin, mupirocin, and antiseptic
resistant S.aureus strains.
Materials and Methods.
This cross-sectional study was performed on a total of 120 MRSA
isolates collected from two major hospitals in Shiraz, Iran. Minimum
inhibitory concentrations (MICs) of vancomycin and mupirocin were
determined by E-test method according to CLSI and Eucast guidelines.
Presence of resistance genes was investigated by PCR method.
Results. Antibacterial susceptibility tests for MRSA isolates showed that three isolates (2.5%) were vancomycin-intermediate S.aureus (VISA), seven isolates (5.8%) were vancomycin-resistant S.aureus
(VRSA), and 15 isolates (12.5%) were high-level mupirocin-resistant
(MuH). None of the isolates had vancomycin resistance gene (vanA), but the frequency of mupirocin resistance gene was significant, and 55 (45.8%) isolates carried the mupA gene. Moreover, norA, smr and qacA/B genes were detected in 110 (91.7%), 55 (45.8%) and 36 (30%) strains, respectively.
Conclusion.
This study showed the existence of VISA and VRSA strains in our region,
and we also found a high frequency of mupirocin and biocide resistance
genes among them.
|
Introduction
Staphylococcus aureus
is an important nosocomial pathogen that can cause superficial and
life-threatening infections. Resistance to antibiotics has made this
organism more problematic.[1] From the 1990s, that
methicillin-resistant Staphylococcus aureus (MRSA) strain has become responsible for one-third of all S. aureus
infections worldwide, and vancomycin is the drug of choice.[1] However,
its increased usage has led to the surge of glycopeptide-resistant S. aureus,
VISA and VRSA, and the resistance mechanisms which have been identified
for VISA and VRSA strains are quite different, which are not fully
understood. For VISA strains, thickening of the bacterial cell wall is
the proposed mechanism of resistance, and for the VRSA strain acquiring
vanA gene from Enterococcus spp.
The worldwide increase in antimicrobial resistance in S. aureus strains
has led to increased mortality and morbidity in human, which highlights
the importance of infection control practices. With this respect,
mupirocin and biocides are increasingly being used in healthcare
systems to eradicate MRSA in individuals who carry it. However, this
increasing usage has led to the occurrence of microorganisms with
reduced susceptibility to them.
Mupirocin (pseudomonic acid A) is
an effective topical antibiotic which is widely used to eliminate MRSA
strains among patients and healthcare workers and is a part of a
comprehensive infection control program to reduce the risk of infection
among the patients who are high risk MRSA carriers.[2] Moreover, it has
been used to control the widespread of MRSA strains among patients
during outbreaks. Another intervention strategy used in clinical
practice to prevent the spread of nosocomial infections is the use of
biocides (including disinfectants and antiseptics), which play a major
role in controlling and preventing nosocomial infections. A wide
variety of biocidal agents, including quaternary ammonium compounds
(QACs), such as benzalkonium chloride and benzethonium chloride and
divalent cations like chlorhexidine digluconate are commonly used in
hospitals and healthcare facilities. In staphylococci, at least 12
biocide resistance genes have been identified: qacA - qacJ, smr and norA.[3]
These determinants encode multidrug resistance efflux pumps that can
mediate reduced susceptibility to either antibiotics or biocides. In S. aureus, qacA, qacB, smr and norA, which encode multidrug-transporter proteins, have been identified as antiseptic-resistance genes.[4]
Given
the importance of VRSA strains which are life-threatening, the spread
of MuR strains, and the resistance to the most common and important
antiseptics in MRSA strains, we aimed to determine the molecular
characteristics of vancomycin, mupirocin and antiseptic resistant S. aureus strains obtained from two teaching hospitals affiliated to Shiraz University of Medical Sciences.
Materials and Methods
Bacterial isolates.
In the present study, 120 clinical MRSA isolates were collected from
October 2012 to March 2013 from two teaching hospitals in Shiraz, Iran.
Outpatient specimens and duplicate isolates were not included. The
isolates were identified as S. aureus
using conventional methods (colony morphology, gram stain, catalase
activity, growth on mannitol salt agar, DNase test, and tube
coagulase).[5] Moreover, all these isolates were investigated for femA and mecA genes for molecular confirmation of MRSA.[6]
Determining the minimum inhibitory concentration.
The MICs of vancomycin (0.016-256 µg/ml) and mupirocin (0.064-1024
µg/ml) were determined by the E-test method on Mueller-Hinton agar
(HiMedia, India) using E-test strips (Liofilchem, Italy) according to
the clinical and laboratory standards institute (CLSI) and Eucast
guidelines.[7,8] Because of the importance of resistance to vancomycin
among the isolates, MICs determination of vancomycin was done twice for
resistant strains. S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 strains were used as the control strains.
Detecting resistance genes.
Bacterial DNA was extracted from the isolates, using the small-scale
phenol-chloroform method, as described previously.[9] Genes encoding
the mupirocin and biocide resistance mupA, qacA/B, smr and norA were
investigated by polymerase chain reaction (PCR), using specific
primers, and the seven isolates that were phenotypically resistant to
vancomycin were evaluated to determine vanA gene.[2,10,11] All of the
mupirocin-resistant isolates that were negative for the mupA gene were investigated for the presence of the mupB
gene. The products were separated by electrophoresis in 1.5% agarose
gels with 1 X TAE (Tris/Acetate/EDTA) buffer, stained with KBC (Kawsar
Biotech Company) load dye and bands were observed by ultraviolet
irradiation.
Results
During
the study period (6 months), MRSA strains were collected from different
infection sources, in which the most were related to the respiratory
tract infection (RTI), 51 isolates (42.5%), followed by skin and soft
tissue infection (SSTI) 22 isolates (18.3%), blood stream infection
(BSI) 19 isolates (15.8%), urinary tract infection (UTI) 15 isolates
(12.5%), sterile fluid infection 7 isolates (5.8%), eye infection 3
isolates (2.5%), and the others 3 isolates (2.5%).
Determining
vancomycin MIC showed that out of 120 studied isolates, 110 (91.7%)
were susceptible; 3 isolates (2.5%) with MIC= 4 µg/ml were intermediate (VISA) and seven isolates (5.2%) were resistant to vancomycin (VRSA) by MIC 256 µg/ml. Resistant strains were evaluated by detecting vanA gene, but all of them were negative for this gene.
By determining mupirocin MIC, with MIC= 8-256 µg/ml the frequency of low-level resistance was 3 (2.5%), and with MIC> 256 µg/ml the high-level resistance was 15 (12.5%). molecular test to determine the frequency of mupA gene
showed a prevalence of 45.8%. The distribution of Low-level mupirocin
resistant (MuL) and High-level mupirocin resistant (MuH) isolates among
different sources of infection is shown in Table 1. Moreover, we found that among MuH strains only seven isolates carried the mupA gene, and the other eight isolates were examined to determine mupB gene, but all of them were negative for this gene.
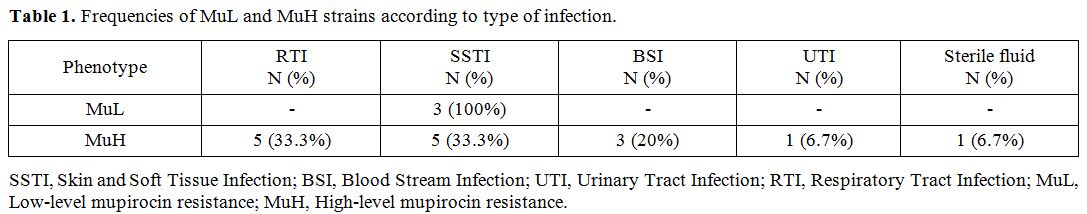 |
Table 1. Frequencies of MuL and MuH strains according to type of infection. |
Results of antiseptic resistant genes revealed that the intended genes, norA, smr and qacA/B were detected in 110 (91.7%), 55 (45.8%) and 36 (30%) MRSA strains, respectively. norA gene was the most frequent resistance genes among MuL and MuH isolates (Table 2). Table 3 shows the co-presence of resistance genes among different sources of infections.
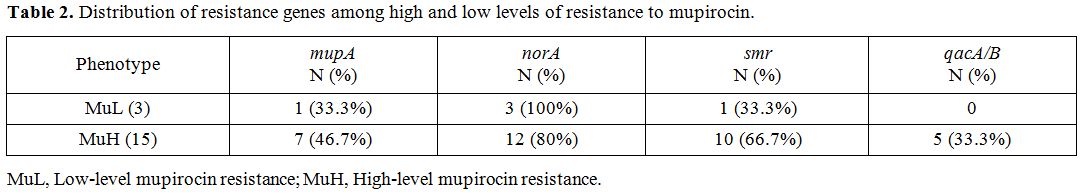 |
Table 2.
Distribution of resistance genes among high and low levels of resistance to mupirocin. |
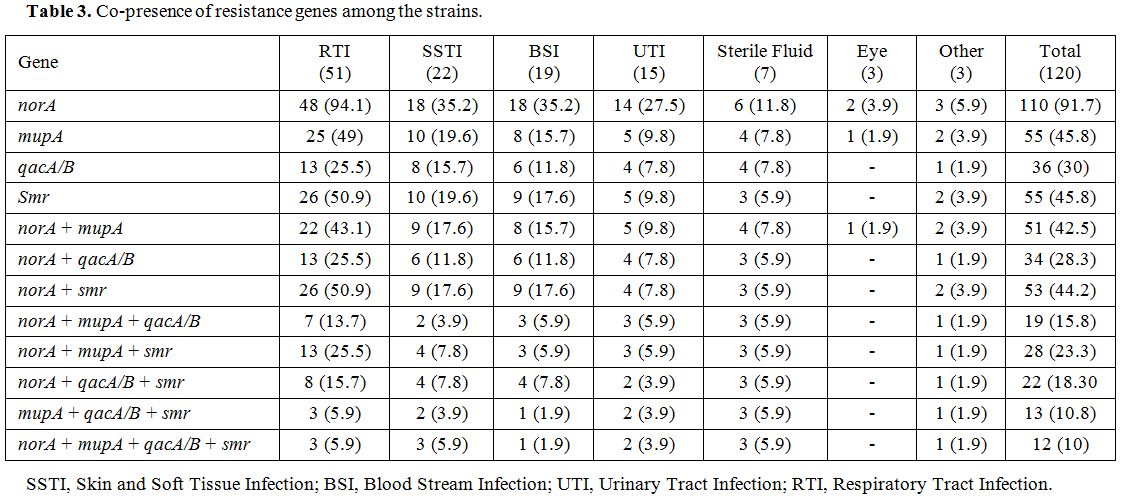 |
Table 3. Co-presence of resistance genes among the strains. |
Discussion
Since the first report of vancomycin-resistant Staphylococcus aureus
(VRSA) strains, many studies have been conducted to determine the
prevalence of these strains.[12] However, to date the rate of strains
with complete resistance to vancomycin (vanA
positive strains) are rare (16 cases from India; 14 cases from the U.S;
6 cases from Iran, and one case in Pakistan), but the frequency of VISA
is relatively high.[13] As the results showed, we found seven strains
that had MIC more than 256 µg/ml and were considered as VRSA. Therefore, we expected to find vanA
gene in these strains as the most common and important cause of
high-level resistance to vancomycin, but in the molecular test, all of
these strains were negative for vanA
gene. Some other studies reported similar cases; for example, in a
study performed by Aligholi et al. in Tehran, Iran among 149 examined
MRSA strains two strains were VRSA strains, one of which was negative
for vanA
gene, but the other was positive.[14] In another study carried out in
Tehran in 2017, Shekarabi et al. reported 4 VRSA strains, one of which
was negative for vanA
gene and the others harbored this gene.[15] Thati et al. reported that
among all VRSA strains, only one was negative for vanA gene.[16] In
Tiwari et al.'s study, out of 783 examined strains in their research,
two strains were found to be vancomycin resistant with no trace of vanA gene.[17] It is supposed that in the absence of vanA
gene the resistance might be expressed through other mechanisms, such
as increase in cell wall thickness based on “vancomycin trapping”
theory, which states that the production of large amounts of
peptidoglycan layers can cause the vancomycin molecules to intercept
the monomers and peptidoglycan layers; as a result, antibiotics are
suppressed before they reach the cell membrane, where the cell wall
synthesis occurs and cannot apply its effect.[17] In addition to the
vancomycin trapping theory, another theory called “affinity trapping”
was presented by Hiramatsu, which states that accumulation of
vancomycin molecules in a thickened wall greatly delays the time to
completely inhibit the cell wall synthesis by preventing sufficient
penetration of vancomycin molecules through thickening of the cell wall
layers.[17] However, in the present study no vanA gene was found amongst the detected VRSA strains, but to date, six cases of vanA
positive VRSA strains have been reported from other parts of Iran, and
according to the latest information, 17 VISA strains have been reported
in addition to the present study.[14,15,18-20] By comparing the
frequency of these strains present in our work and worldwide
literature, there is evidence that our country is at risk of increasing
resistance to this critical antibiotic. Hence, more attention to
managing the resistance process is necessary, especially for VISA
strains, associated with persistent infections, poor clinical outcomes,
prolonged vancomycin treatment or treatment failure.Furthermore,
we investigated different indices on the seven VRSA isolates and found
that most isolates were isolated from respiratory tract infections
(42.8%), followed by bloodstream infections (28.5%). Also, we found
that all VRSA strains are of high-level resistance to mupirocin, except
for one other isolate which carries the norA gene (Table 4).
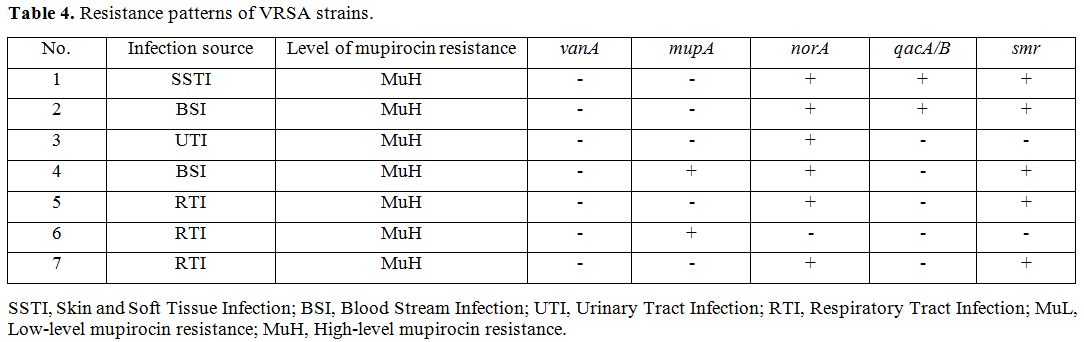 |
Table 4. Resistance patterns of VRSA strains. |
As
mentioned previously, increased use of mupirocin has led to the
occurrence of mupirocin resistance strain which is considered as a
significant alarm. Hence, it is crucial to determine the frequency and
distribution of these resistant strains. In the present study, we found
18 mupirocin-resistant strains, 15 of which showed MuH phenotype. The
molecular test showed that among 15 MuH strains, only seven strains
carried mupA gene and the eight others lacked this gene. Since the mupB
gene was discussed recently as another contributing factor in
resistance to mupirocin,[2] we also considered the possibility of the
presence of this gene, but all the isolates were negative for the mupB gene. Actually, we examined all the isolates for the presence of the mupA gene, and 55 of them (45.8%) carried mupA gene.The
results of the present study showed that resistance to mupirocin
(12.5%) in our region has increased since in the previous scientific
literature no resistance to mupirocin was reported.[21] This frequency
was high in comparison with other reports in the country. In the study
performed in Tehran, the incidence of MuL strains was 3.5%, MuH was 1%,
and mupA gene was 5.8%.[22] A study in Arak reported that the frequency of MuH was 7.3%, and that of the mupA gene
was 6%.[23] Goudarzi et al. reported among burn patients with
bacteremia in Teheran that the frequency of MuH was 19.8% and 6.5%,
respectively.[24,25] However, the results of our study are in line with
those of Nepal in which the frequency of mupA
gene was 48.3%, but the prevalence of MuH strains was 51%.
Consequently, it is suggested that this significant difference might be
due to the pattern of consuming this antibiotic, different geographic
regions, and the study population. According to these results and the
increase in antibiotic consumption that causes resistance, it is
necessary to pay more attention to the manner and rate of using
mupirocin to prevent further resistance.The frequency of norA gene,
as a biocide resistance gene, in these strains was notable and all
(100%) of MuL strains and 80% of MuH isolates harbored it. norA
encoded efflux pumps can create resistance either to biocides or
antibiotics. Resistance to mupirocin might be related to such efflux
pumps. Moreover, out of 55 isolates carrying mupA,
48 were susceptible to mupirocin; cases like this have been
reported,[26] and the probable cause has been announced to be lack of
gene expression. The emergence of antibiotics resistant S. aureus
has made it difficult to treat and control the infections caused by
this organism. Also, this ability gives them the potential to stabilize
in healthcare settings as well as their transmission among patients.
Consequently, prevention and control have reduced the presence of these
problematic strains through appropriate measures and proper use of
proper disinfectants. Nonetheless, studies have shown that prolonged
and inappropriate use of disinfectants can lead to the development of
resistant strains.[27] The most abundant and critical anti-disinfectant
determinants studied are qacA/B, smr and norA genes.[28] In the present study, the frequencies of norA, smr and qacA/B genes were 91.7%, 45.8%, and 30%, respectively. The frequency of norA gene was the highest, and that of smr was higher than qacA/B, which was similar to a previous study.[29] The rate of qacA/B
gene in different countries ranges from 2% in Canada to 61% to 83% in
China, and that of smr gene varies from 37.4% to 59% in China. In fact,
to date, these genes have been reported most frequently in China.[10]
Many other studies, as well as ours, reported the presence of both
genes together,[10,30-32] where the prevalence of these strains was
19.1%.
Conclusions
MIC
tests of vancomycin showed that 91.7% of isolates were susceptible. The
frequencies of VISA and VRSA were 2.5% and 5.2%, respectively. vanA
gene was not detected in any of resistant strains. Mupirocin MIC tests
showed that the frequency of low-level resistance (MuL) was 2.5% and
high-level resistance (MuH) was 12.5%. The distribution of mupA gene was 45.8%. Only seven MuH isolates carried mupA gene, and all of them were negative for mupB gene amplification. Investigation of antiseptic resistant genes revealed that norA, smr and qacA/B
were detected in 91.7%, 45.8% and 30% of MRSA strains, respectively.
According to the data, more attention is required when using these
antibiotics, and antiseptics are suggested to control the range of
resistance.
References
- Samanta, D. and Elasri, M.O. The msaABCR operon
regulates resistance in vancomycin-intermediate Staphylococcus aureus
strains. Antimicrob. Agents Chemother. 2014; 58, 6685-95. https://doi.org/10.1128/AAC.03280-14 PMid:25155591 PMCid:PMC4249426

- Seah,
C., Alexander, D.C., Louie, L., Simor, A., Low, D.E., Longtin, J. and
Melano, R.G. MupB, a new high-level mupirocin resistance mechanism in
Staphylococcus aureus. Antimicrob Agents Chemother. 2012; 56, 1916-20. https://doi.org/10.1128/AAC.05325-11 PMid:22252810 PMCid:PMC3318397

- Vali,
L., Davies, S.E., Lai, L.L., Dave, J. and Amyes, S.G. Frequency of
biocide resistance genes, antibiotic resistance and the effect of
chlorhexidine exposure on clinical methicillin-resistant Staphylococcus
aureus isolates. J Antimicrob Chemother. 2008; 61, 524-32. https://doi.org/10.1093/jac/dkm520 PMid:18227090

- Noguchi,
N., Suwa, J., Narui, K., Sasatsu, M., Ito, T., Hiramatsu, K. and Song,
J.H. Susceptibilities to antiseptic agents and distribution of
antiseptic-resistance genes qacA/B and smr of methicillin-resistant
Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med
Microbiol. 2005; 54, 557-65. https://doi.org/10.1099/jmm.0.45902-0 PMid:15888465

- Kloss
WE, B.T. Staphylococcus and Micrococcus. Manual of Clinical
Microbiology, ed. B.E. Murray PR, Pfaller MA, Tenover FC and Yolken
RH1995, Washington DC: ASM. 282-284.
- Kobayashi,
N., Wu, H., Kojima, K., Taniguchi, K., Urasawa, S., Uehara, N., Omizu,
Y., Kishi, Y., Yagihashi, A. and Kurokawa, I. Detection of mecA, femA,
and femB genes in clinical strains of staphylococci using polymerase
chain reaction. Epidemiol Infect. 1994; 113, 259-66. https://doi.org/10.1017/S0950268800051682 PMid:7925664 PMCid:PMC2271538

- Clinical
and Laboratory Standards Institute, Performance Standards for
Antimicrobial.Susceptibility Testing; Twenty-sixth Informational
Supplemen, 2016 (m100-s26).
- The
European Committee on Antimicrobial Susceptibility Testing. Breakpoint
tables for interpretation of MICs and zone diameters [Internet].
Breakpoint tables for interpretation of MICs and zone diameters; 2016.
- Sambrook,
J. and Russell, D.W. Purification of nucleic acids by extraction with
phenol: chloroform. Cold Spring Harbor Protocols. 2006; 4455. https://doi.org/10.1101/pdb.prot4455 PMid:22485786

- Liu,
Q., Zhao, H., Han, L., Shu, W., Wu, Q. and Ni, Y. Frequency of
biocide-resistant genes and susceptibility to chlorhexidine in
high-level mupirocin-resistant, methicillin-resistant Staphylococcus
aureus (MuH MRSA). Diagn Microbiol Infect Dis. 2015; 82, 278-83. https://doi.org/10.1016/j.diagmicrobio.2015.03.023 PMid:26008124

- Clark,
N.C., Cooksey, R.C., Hill, B.C., Swenson, J.M. and Tenover, F.C.
Characterization of glycopeptide-resistant enterococci from US
hospitals. Antimicrob. Agents Chemother. 1993; 37, 2311-17. https://doi.org/10.1128/AAC.37.11.2311 PMid:8285611 PMCid:PMC192384

- Hiramatsu,
K., Aritaka, N., Hanaki, H., Kawasaki, S., Hosoda, Y., Hori, S.,
Fukuchi, Y. and Kobayashi, I. Dissemination in Japanese hospitals of
strains of Staphylococcus aureus heterogeneously resistant to
vancomycin. The Lancet. 1997; 350, 1670-73. https://doi.org/10.1016/S0140-6736(97)07324-8

- McGuinness,
W.A., N. Malachowa, and F.R. DeLeo. Vancomycin Resistance in
Staphylococcus aureus. Yale J Biol Med. 2017; 90, 269-81. PMid:28656013
PMCid:PMC5482303

- Aligholi,
M., Emaneini, M., Jabalameli, F., Shahsavan, S., Dabiri, H. and
Sedaght, H. Emergence of high-level vancomycin-resistant Staphylococcus
aureus in the Imam Khomeini Hospital in Tehran. Med Princ Pract. 2008;
17, 432-4. https://doi.org/10.1159/000141513 PMid:18685289

- Shekarabi,
M., Hajikhani, B., Chirani, A.S., Fazeli, M. and Goudarzi, M. Molecular
characterization of vancomycin-resistant Staphylococcus aureus strains
isolated from clinical samples: A three year study in Tehran, Iran.
PLoS One. 2017; 12, e0183607. https://doi.org/10.1371/journal.pone.0183607 PMid:28854219 PMCid:PMC5576738

- Thati,
V., C.T. Shivannavar, and S.M. Gaddad. Vancomycin resistance among
methicillin resistant Staphylococcus aureus isolates from intensive
care units of tertiary care hospitals in Hyderabad. Indian J Med Res.
2011; 134, 704-8. https://doi.org/10.4103/0971-5916.91001 PMid:22199111 PMCid:PMC3249970

- Tiwari,
H.K. and M.R. Sen. Emergence of vancomycin resistant Staphylococcus
aureus (VRSA) from a tertiary care hospital from northern part of
India. BMC Infect Dis. 2006; 6, 156. https://doi.org/10.1186/1471-2334-6-156 PMid:17067393 PMCid:PMC1634751

- Dezfulian,
A., Aslani, M.M., Oskoui, M., Farrokh, P., Azimirad, M., Dabiri, H.,
Salehian, M.T. and Zali, M.R. Identification and characterization of a
high vancomycin-resistant Staphylococcus aureus harboring vanA gene
cluster isolated from diabetic foot ulcer. Iran J Basic Med Sci. 2012;
15, 803-6. PMid:23495359 PMCid:PMC3586877

- Azimian,
A., Havaei, S.A., Fazeli, H., Naderi, M., Ghazvini, K., Samiee, S.M.,
Soleimani, M. and Peerayeh, S.N. Genetic characterization of a
vancomycin-resistant Staphylococcus aureus isolate from the respiratory
tract of a patient in a university hospital in northeastern Iran. J
Clin Microbiol. 2012; 50, 3581-5. https://doi.org/10.1128/JCM.01727-12 PMid:22933598 PMCid:PMC3486207

- Hasani,
A., Sheikhalizadeh, V., Hasani, A., Naghili, B., Valizadeh, V. and
Nikoonijad, A.R. Methicillin resistant and susceptible Staphylococcus
aureus: Appraising therapeutic approaches in the Northwest of Iran.
Iran J Microbiol. 2013; 5, 56-62. PMid:23467268 PMCid:PMC3577566

- Askarian,
M., Zeinalzadeh, A., Japoni, A., Alborzi, A. and Memish, Z.A.
Prevalence of nasal carriage of methicillin-resistant Staphylococcus
aureus and its antibiotic susceptibility pattern in healthcare workers
at Namazi Hospital, Shiraz, Iran. Int J Infect Dis. 2009; 13, 241-47. https://doi.org/10.1016/j.ijid.2008.11.026 PMid:19269873

- Saderi,
H., Oulia, P. and Habibi, M. Detection of resistance to mupirocin in
Staphylococcus aureus strains isolated from patients in four university
hospitals of Tehran by polymerase chain reaction (pcr) method.
Daneshvar Medicine, December 2008-January 2009, Volume 16 , Number 78;
29-36.

- Hesami,
S., Hosseini, S.D., Amouzandeh-Nobaveh, A., Eskandari, S. and
Ghaznavi-Rad, E. Phenotypic and genotypic determination of mupirocin
resistance among methicillin susceptibility and resistance in
Staphylococci isolated from nosocomial infections. Journal of
Mazandaran University of Medical Sciences. 2014; 23, 30-39.

- Goudarzi,
M., Seyedjavadi, S.S., Nasiri, M.J., Goudarzi, H., Nia, R.S. and
Dabiri, H. Molecular characteristics of methicillin-resistant
Staphylococcus aureus (MRSA) strains isolated from patients with
bacteremia based on MLST, SCCmec, spa, and agr locus types analysis.
Microb Pathog. 2017; 104, 328-35. https://doi.org/10.1016/j.micpath.2017.01.055 PMid:28159661

- Goudarzi,
M., Bahramian, M., Tabrizi, M.S., Udo, E.E., Figueiredo, A.M.S.,
Fazeli, M. and Goudarzi, H. Genetic diversity of methicillin resistant
Staphylococcus aureus strains isolated from burn patients in Iran:
ST239-SCCmec III/t037 emerges as the major clone. Microb Pathog. 2017;
105, 1-7. https://doi.org/10.1016/j.micpath.2017.02.004 PMid:28179118

- Fritz,
S.A., Hogan, P.G., Camins, B.C., Ainsworth, A.J., Patrick, C., Martin,
M.S., Krauss, M.J., Rodriguez, M. and Carey-Ann, B.D. Mupirocin and
chlorhexidine resistance in Staphylococcus aureus in patients with
community-onset skin and soft tissue infections. Antimicrob. Agents
Chemother. 2013; 57, 559-68. https://doi.org/10.1128/AAC.01633-12 PMid:23147738 PMCid:PMC3535967

- McDonnell,
G. and A.D. Russell. Antiseptics and disinfectants: activity, action,
and resistance. Clin Microbiol Rev. 1999; 12, 147-79. PMid:9880479
PMCid:PMC88911

- Jaglic,
Z. and D. Cervinkova. Genetic basis of resistance to quaternary
ammonium compounds--the qac genes and their role: a review. Veterinarni
Medicina. 2012; 57. https://doi.org/10.17221/6013-VETMED

- Nowroozi,
J., Pakzad, P., Ebrahimi, E. and Razavipour, R. Detection of biocide
resistance genes, qacA/B and smr, among isolated Staphylococcus aureus
from clinical and non-clinical sources. Pajoohandeh Journal. 2011; 16,
83-91.

- Mayer,
S., Boos, M., Beyer, A., Fluit, A.C. and Schmitz, F.J. Distribution of
the antiseptic resistance genes qacA, qacB and qacC in 497
methicillin-resistant and -susceptible European isolates of
Staphylococcus aureus. J Antimicrob Chemother. 2001; 47, 896-7. https://doi.org/10.1093/jac/47.6.896 PMid:11389128

- Noguchi,
N., Suwa, J., Narui, K., Sasatsu, M., Ito, T., Hiramatsu, K. and Song,
J.H. Susceptibilities to antiseptic agents and distribution of
antiseptic-resistance genes qacA/B and smr of methicillin-resistant
Staphylococcus aureus isolated in Asia during 1998 and 1999. J Med
Microbiol. 2005; 54, 557-65. https://doi.org/10.1099/jmm.0.45902-0 PMid:15888465

- Taheri,
N., Ardebili, A., Amouzandeh-Nobaveh, A. and Ghaznavi-Rad, E. Frequency
of antiseptic resistance among Staphylococcus aureus and
coagulase-negative staphylococci isolated from a university hospital in
central Iran. Oman medical journal. 2016; 31, 426. https://doi.org/10.5001/omj.2016.86 PMid:27974958 PMCid:PMC5099399

[TOP]


































