Fatma Devrim, Erkin
Serdaroğlu, İlknur Çağlar, Yeliz Oruç, Nevbahar Demiray, Nuri Bayram,
Hasan Ağın, Sebnem Çalkavur, Yelda Sorguç, Nida Dinçel, Yüce Ayhan,
Ebru Yılmaz and Ilker Devrim.
Dr. Behçet Uz Children's Hospital, İzmir, Turkey
Correspondence to: Nuri Bayram, Medical Doctor. Dr. Behçet Uz
Children's Hospital, İzmir, Turkey. Tel: +90 232 4116196, Fax: +90 232
4892315. E-mail:
nuribayram@gmail.com
Published: September 1, 2018
Received: May 18, 2018
Accepted: August 3, 2018
Mediterr J Hematol Infect Dis 2018, 10(1): e2018055 DOI
10.4084/MJHID.2018.055
This article is available on PDF format at:

This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract Background. Healthcare-associated
infections results in increased health care costs and mortality. There
are limited studies concerning the distribution of the etiologic agents
and the resistance patterns of the microorganisms causing
healthcare-associated urinary tract infections (HA-UTI) in pediatric
settings.
Objectives. The
aim of this study was to evaluate the distribution and antibiotic
susceptibility patterns of pathogens causing HA-UTI in children.
Material and Methods.
Isolates from 138 children with UTI who were hospitalized in pediatric,
neonatal and pediatric surgery intensive care units were reviewed.
Results.
Most common isolated organism was Klebsiella pneumoniae (34.1%) and
Escherichia coli (26.8%). Among the Pseudomonas aeruginosa, Meropenem
and imipenem resistance rates were 46.2% and 38.5%. Extended-spectrum
beta-lactamase (ESBL) production was present in 48 Klebsiella species
(82.8%). Among ESBL positive Klebsiella species, the rate of meropenem
and imipenem resistance was 18.8%, and ertapenem resistance was 45.9%.
Extended spectrum beta-lactamase production was present in 27 (72.9%)
Escherichia coli species. Among ESBL positive E. coli, the rate of
meropenem and imipenem resistance was 7.4%, and ertapenem resistance
was 14.8%
Conclusions.
Emerging meropenem resistance in P. aeruginosa, higher rates of
ertapenem resistance in ESBL positive ones in E. coli and Klebsiella
species in pediatric nosocomial UTI are important notifying signs for
superbug infections.
|
Introduction
Healthcare-associated
infections (HAIs) are common and probably one of the most preventable
complications during hospitalization resulting in increased health care
costs and mortality.[1] According to CDC, healthcare-associated urinary
tract infections (HA-UTIs) in the United States acute care hospitals
are estimated to be about 93 300 annually in 2011.[2] Urinary tract
infection was reported to be leading HAI among hospitalized adults and
in critical care units[3,4] and the second or third most common type of
nosocomial infection in intensive care units (ICUs).[5-7] The HA-UTI is
frequently related to bladder catheterization,[3,4] and the risk of
catheter-associated urinary tract infection (CA-UTI) is reported to
increase by 3% to 7% within each day of the indwelling urinary catheter
remains.[8,9]
Most epidemiological data including the
distribution of the etiologic agents and the resistance patterns of the
microorganisms causing HA-UTI is mainly based on adult reports, and
there are limited studies concerning the isolated HA-UTI in
children.[4,10] In addition, most of the studies about nosocomial UTIs
are mostly related to CA-UTI. Therefore the real epidemiology of
symptomatic non-catheter-associated UTI (Non-CAUTI) has not been
established in the pediatric settings. Thus, the objective of the study
is to evaluate the distribution and also the antibiotic susceptibility
patterns of pathogens causing HA-UTI, with especially focusing on
whether it is catheter-associated or non-catheter-associated UTI in
children hospitalized at ICUs.
Materials and Methods
Study subjects and methods.
This study included the symptomatic HA-UTI in children under 18 years
old who were hospitalized in the ICUs of Dr. Behçet Uz Children’s
Hospital between the periods from January 2014 to December 2017. This
hospital is a referral center for pediatric patients in the Aegean
Region of Turkey with annual outpatient 600 000 patients and
approximately 23 000 hospitalizations in 2016. The pediatric intensive
care unit (PICU) has 24-bed capacity with 500 hospitalizations, the
neonatal intensive care unit (NICU) has 60 bed-capacities with 1500
hospitalizations, and the pediatric surgery and the pediatric
cardiovascular surgery ICUs have 6–bed capacities and 200
hospitalizations, annually.
Definitions.
All children in ICUs who were diagnosed as symptomatic UTI with
positive urinary culture results were included to study. The
definitions of symptomatic UTI including symptomatic CA-UTI and
non-CAUTI were defined according to the definitions of Centers for
Disease Control and Prevention.[11]
Microbiological analysis.
Each urinary culture was placed in the Bac T/ALERT 9240 95 automated
system (bioMérieux, Marcy l’Etoile, France) and incubated for seven
days or until they were found to be positive.[12] The microorganisms
were identified with the VITEK-2 compact system (bioMérieux), and
antibiotic susceptibility tests (including MIC levels, ESBL presence,
and carbapenem resistance) were also performed with the same system for
each isolate according to the manufacturer’s instructions and the
European Committee on Antimicrobial Susceptibility Testing.
Identification and antibiotic susceptibility tests of gram-positive
bacteria were performed using the automated VITEK-2 system with
gram-positive identification card AST-P592, a supplementary E-test
(bioMérieux, Durham, NC, USA), and a disk diffusion test according to
the manufacturer’s instructions. Vancomycin-resistant Enterococcus spp. (VRE) and methicillin-resistant Staphylococcus aureus
(MRSA) were also identified using the automated VITEK-2 system.[13]
This system was also used for the identification and antibiotic
susceptibility tests of gram-negative bacteria with Gram-negative
identification card AST-N325, AST-N326, and AST-N327.[14]
This study was approved by the Local Ethical Committee of 120 Dr. Behçet Uz Children’s Training and Research Hospital.
Statistical analysis.
Statistical analysis was performed using SPSS, version 15.0 1 (IBM
SPSS, Chicago, IL). Quantitative data are expressed as a mean and
standard deviation or median with interquartile range (IQR) if data
followed a non-normal distribution. Qualitative variables were
expressed as absolute and relative frequencies. Chi-square, with
Fisher’s exact correction where required for discrete variables,
and Student’s t-test for parametric and Wilcoxon rank sum test for
non-parametric continuous variables were used. Probabilities (p values)
less than 0.05 were considered significant for all tests.
Results
During
the study period, a total of 152 nosocomial symptomatic UTI episodes
were recorded. Fourteen of these were excluded due to absent data. A
total of 138 UTI episodes which had complete medical files and
susceptibility patterns were included in the final analysis. Among the
138 UTI episodes, 74 (53.6%) episodes were in NICU, 55 (39.9%) episodes
were in PICU, 7 (5.1%) episodes were in pediatric surgery ICU, and 2
(1.4%) episodes were in pediatric cardiovascular surgery ICU. Of all
analyzed UTIs, 26 (18.8%) were symptomatic CA-UTI and 112 (81.2%) were
symptomatic non-CAUTI.
Gram-negative microorganisms were the most
common isolated organisms (119 isolations, 86.2%) followed by
Gram-positive bacteria (13 isolations, 9.4%) and Candida spp. (4.3%). The distribution of the isolated microorganisms was reviewed in Table 1. The most common isolated organism was K. pneumoniae (34.1%) and E. coli (26.8%) followed by other microorganisms reviewed in Table 1.
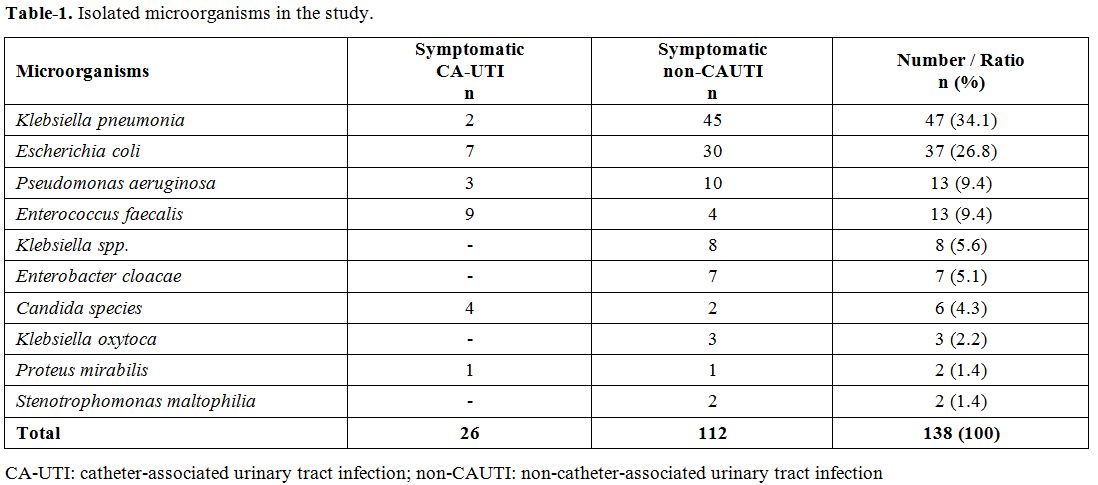 |
Table 1. Isolated microorganisms in the study. |
Resistance patterns. Among the P. aeruginosa,
in vitro susceptibility was highest to amikacin, followed by colistin,
gentamicin, tobramycin, levofloxacin, and ciprofloxacin. Meropenem and
imipenem resistance rates were 46.2% and 38.5%, consecutively (Table 2). Nearly 53.8% of the P. aeruginosa were resistant to ceftazidime showing the highest resistance rate.
Extended-spectrum beta-lactamase (ESBL) production was present in 48 Klebsiella species (82.8%). Among ESBL positive Klebsiella species, the rate of meropenem and imipenem resistance was 18.8%, and ertapenem resistance was 45.9% (Table 2). Aminoglycoside resistance ranges from 8.3 to 43.8% in Klebsiella
species, and ciprofloxacin resistance were present in 39.6% of the
isolates. Colistin resistance was observed in 12.5% of the Klebsiella species isolate (Table 2).
Extended spectrum beta-lactamase production was present in 27 (72.9%) of E. coli species. Among ESBL positive E. coli, the rate of meropenem and imipenem resistance was 7.4%, and ertapenem resistance was 14.8% (Table 2).
Aminoglycoside resistance ranged from 25.9% to 66.7% (amikacin,
tobramycin, and gentamicin resistance were 25.9%, 66.7%, 66.7%,
respectively) and ciprofloxacin resistance was present in 33.3% in ESBL
positive E. coli species. Resistance to colistin was not observed in E. coli isolates.
Among 7 Enterobacter cloaca
strains, only 1 (14.3%) was ESBL positive, and this isolate was
resistant to meropenem, imipenem, aminoglycosides, and other
antimicrobial agents.
Among the 13 Enterococcus fecalis strains, vancomycin resistance was present in 2 isolates (15.4%), while all isolates were susceptible to linezolid.
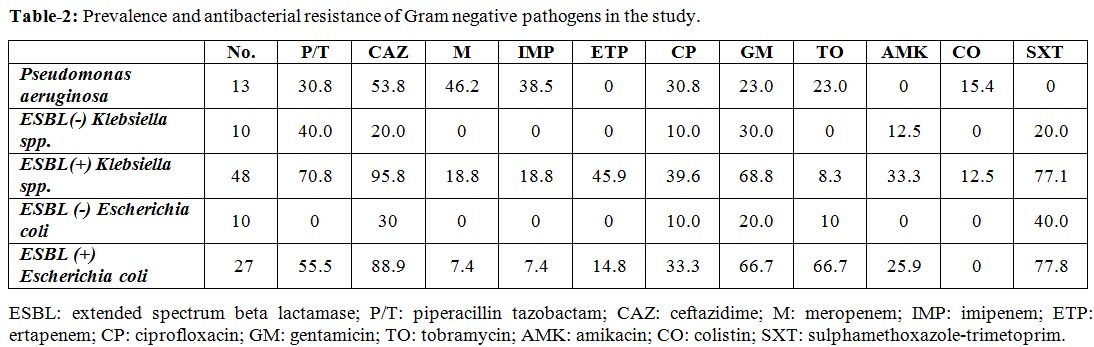 |
Table
2. Prevalence and antibacterial resistance of Gram negative pathogens in the study. |
Discussion
In
this cross-sectional study, the pathogens causing symptomatic HA-UTIs
and their resistance patterns are evaluated. The most common isolated
species were Klebsiella spp. followed by E. coli and P. aeruginosa isolates. Extended spectrum beta-lactamase production was present in 82.8% of the 48 Klebsiella species and 72.9% of the E. coli species. Among ESBL positive E. coli and Klebsiella
species, the rate of meropenem (imipenem) resistance was 18.8% and 7.4%
while ertapenem resistance was found to be higher and 45.9% in Klebsiella species and 14.8 in E. coli species.
Although the dominant pathogen in children was reported to be E. coli[15-20] in previous studies, Klebsiella
species were the most common isolated organisms as HA-UTI pathogen in
the current study. In a study of European Study Group on Nosocomial
infections group including 298 patients, E. coli (35.3%) was the most commonly isolated organism, and Klebsiella spp. were reported as 9.8% of the pathogens.[20]
Emerging
of resistance among uropathogens is increasingly reported within a
variety of resistant patterns.[21,22] In this study, the rate of ESBL
positive Klebsiella
species was 82.8%, and meropenem resistance was 18.8%, while ertapenem
resistance was reported to be 45.9%. In one study from our center which
had focused on 335 ESBL-producing Enterobacteriaceae including 193
urinary tract pathogens, meropenem resistance was not reported, and
ertapenem resistance was reported to be as low as 8.5% in 2009.[23]
Although this was a cross-sectional comparison, a remarkable increase
in resistance patterns for Klebsiella
species was observed. As well as in other studies worldwide,[24-26]
there is an undesirable trend toward the emergence of carbapenemase
resistance. Since its first detection in 1996, carbapenemase-producing Klebsiella pneumoniae (KPC) had been an important medical problem, and the rate of KPC production was high enough to have serious concern.[24]
Although ESBL production was observed in 72.9% of the E. coli isolates in this study, the rate of carbapenem resistance was much lower compared to Klebsiella
species. In one study from India reported a dramatic increase over the
5-year study period.[21] İlker et al. reported that 99% of the
ESBL-producing E. coli
isolates in their center during the period of 2009, was found to be
susceptible to ertapenem and 100% to meropenem, however the ertapenem
resistance increased to 14.8% and meropenem to 7.4% suggesting the
emerging resistance during the last five years.[23]
The rates of resistance to aminoglycoside have a wide spectrum ranging from 8.3% to 43.8% in Klebsiella species and from 6% to 66.7% in ESBL positive E. coli isolates. Resistance to colistin in E. coli isolates
was not observed. The use of amikacin monotherapy for UTI with
ESBL-producing bacteria in children is limited, and Polat et al.
reported that this treatment regimen might be a reasonable
alternative.[27] In our study, the resistance patterns suggested that
the selection of the type of aminoglycoside is also important due to
different resistance patterns.
This study has some limitations due
to its retrospective design. The study included resistance patterns of
the common pathogens of nosocomial UTI and did not focus on mortality
and treatment response. Secondly, the timeline trends of resistance
patterns for specific bacteria were not compared due to the
cross-section pattern, while the data including current study was
compared to the previous study from our center in 2009.
Most
epidemiological data on nosocomial resistance patterns are limited to
adult studies and generally focused on studies about nosocomial CA-UTI.
In our study emerging meropenem resistance in P. aeruginosa, ESBL production and higher rates of ertapenem resistance in ESBL positive ones in E. coli and Klebsiella
species, in nosocomial UTI are important notifying signs for the
development of superbug infections also in children in the future.
References
- World Health Organization. Prevention of Hospital-Acquired Infections. 2nd edn. Geneva: WHO; Available from: www.who.int/csr/resources/publications/whocdscsreph200212.pdf, 2002. (last check: January 15, 2018).

- CDC. Healthcare-associated Infections(HAI). HAI Data and Statistics https://www.cdc.gov/hai/surveillance/index.html (last check: January 15, 2018).
- Haley
RW, Culver DH, White JW, Morgan WM, Emori TG. The Nationwide Nosocomial
Infection Rate: a new need for vital statistics. Am J Epidemiol.
1985;121:159-67. https://doi.org/10.1093/oxfordjournals.aje.a113988
PMid:4014113

- Laupland
KB, Zygun DA, Davies HD, Church DL, Louie TJ, Doig CJ. Incidence and
risk factors for acquiring nosocomial urinary tract infection in the
critically ill. J Crit Care. 2002;17:50-7.
https://doi.org/10.1053/jcrc.2002.33029 PMid:12040549

- Rosenthal
VD, Maki DG, Jamulitrat S, Medeiros EA, Kumar TS, Yepes Gómez D, et al.
International Nosocomial Infection Con- trol Consortium (INICC) report,
data summary for 2003-2008, issued June 2009. Am J Infect Control.
2010;38:95-106. https://doi.org/10.1016/j.ajic.2009.12.004
PMid:20176284

- National
Nosocomial Infections Surveillance (NNIS) System Report: data summary
from January 1992 trough June 2004, issued October 2004. Am J infect
Control. 2004; 32:470-85. https://doi.org/10.1016/j.ajic.2004.10.001
PMid:15573054

- Bustinza
Arriortúa A, Solana García MJ, Botrán Prieto M, Padilla Ortega B.
Infección nosocomial. In: Manual de Cuida dos Intensivos Pediátricos.
3a ed. Madrid: Publimed; 2009. p. 323-35.
- McGuckin
M. The patient survival guide: 8 simple solutions to prevent hospital
and healthcare-associated infections. New York, NY: Demos Medical
Publishing; 2012.
- Lo E, Nicolle LE,
Coffin SE, Gould C, Maragakis LL, Meddings J, et al. Strategies to
prevent catheter-associated urinary tract infections in acute care
hospitals: 2014 update. Infect Control Hosp Epidemiol. 2014;35:464-79.
https://doi.org/10.1086/675718 PMid:24709715

- McGregor
JC, Quach Y, Bearden DT, Smith DH, Sharp SE, Guzman-Cottrill JA.
Variation in antibiotic susceptibility of uropathogens by age among
ambulatory pediatric patients. J Pediatr Nurs. 2014;29:152-7.
https://doi.org/10.1016/j.pedn.2013.09.001 PMid:24091131
PMCid:PMC3943820

- CDC
https://www.cdc.gov/nhsn/pdfs/pscmanual/7psccauticurrent.pdf
Urinary Tract Infection (Catheter-Associated Urinary Tract Infection
[CAUTI] and Non-Catheter-Associated Urinary Tract Infection [UTI]) and
Other Urinary System Infection [USI]) Events (last updated April 23,
2018)
- Wilson ML, Weinstein MP and Reller
LB. Automated blood culture systems. Clin Lab Med. 1994;14:149–69.
https://doi.org/10.1016/S0272-2712(18)30401-3

- Bobenchik
AM, Hindler JA, Giltner CL, et al. Performance of Vitek 2 for
antimicrobial susceptibility testing of Staphylococcus spp. and
Enterococcus spp. J Clin Microbiol. 2014;52:392–7.
https://doi.org/10.1128/JCM.02432-13 PMid:24478467 PMCid:PMC3911353

- Quesada
MD, Giménez M, Molinos S, et al. Performance of VITEK-2 compact and
overnight MicroScan panels for direct identification and susceptibility
testing of Gram- negative bacilli from positive FAN BacT/ALERT blood
culture bottles. Clin Microbiol Infect. 2010;16:137–40.
https://doi.org/10.1111/j.1469-0691.2009.02907.x PMid:19778301

- Gruneberg
RN. Changes in urinary pathogens and their antibiotic sensitivities,
1971-1992. J Antimicrob Chemother. 1994;33:1-8.
https://doi.org/10.1093/jac/33.suppl_A.1 PMid:7928827

- Gupta
K, Scholes D, Stamm WE. Increasing prevalence of antimicrobial
resistance among uropathogens causing acute uncomplicated cystitis in
women. JAMA. 1999;281:736-738. https://doi.org/10.1001/jama.281.8.736
PMid:10052444

- Prais
D, Straussberg R, Avitzur Y, Nussinovitch M, Harel L, Amir J. Bacterial
susceptibility to oral antibiotics in community acquired urinary tract
infection. Arch Dis Child. 2003;88:215-8.
https://doi.org/10.1136/adc.88.3.215 PMid:12598381 PMCid:PMC1719471

- Ashkenazi
S, Even-Tov S, Samra Z, Dinari G. Uropathogens of various child- hood
populations and their antibiotic susceptibility. Pediatr Infect Dis J.
1991; 10:742-6. https://doi.org/10.1097/00006454-199110000-00005
PMid:1945576

- Lutter
SA, Currie ML, Mitz LB, Greenbaum LA. Antibiotic resistance patterns in
children hospitalized for urinary tract infections. Arch Pediatr
Adolesc Med. 2005;159:924-8.
https://doi.org/10.1001/archpedi.159.10.924 PMid:16203936

- Bouza
E, San Juan R,Mu-oz P,Voss A, Kluytmans J;Co-operative Group of the
European Study Group on Nosocomial Infections. A European perspective
on nosocomial urinary tract infections II. Report on incidence,
clinical characteristics and outcome (ESGNI-004 study). European Study
Group on Nosocomial Infection. Clin Microbiol Infect. 2001;7:532-42.
https://doi.org/10.1046/j.1198-743x.2001.00324.x PMid:11683793

- Patwardhan
V, Kumar D, Goel V, Singh S Changing prevalence and antibiotic drug
resistance pattern of pathogens seen in community-acquired pediatric
urinary tract infections at a tertiary care hospital of North India. J
Lab Physicians. 2017;9:264-8. https://doi.org/10.4103/JLP.JLP_149_16
PMid:28966488 PMCid:PMC5607755

- Bryce
A, Hav AD, Lane IF, Thornton HV, Wootton M, Costelloe C. Global
prevalence of antibiotic resistance in paediatric urinary tract
infections caused by Escherichia coli and association with routine use
of antibiotics in primary care: systematic review and meta-analysis.
BMJ. 2016;352:i939. https://doi.org/10.1136/bmj.i939 PMid:26980184
PMCid:PMC4793155

- Devrim
I, Gulfidan G, Gunay I, Agın H, Güven B, Yılmazer MM, Dizdarer C.
Comparison of in vitro activity of ertapenem with other carbapenems
against extended-spectrum beta-lactamase-producing Escherichia coli and
Klebsiella species isolated in a tertiary children's hospital. Expert
Opin Pharmacother. 2011;12:845-9.
https://doi.org/10.1517/14656566.2011.559460 PMid:21323503

- Yigit
H, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a
carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents
Chemother. 2001;45:1151–61.
https://doi.org/10.1128/AAC.45.4.1151-1161.2001 PMid:11257029
PMCid:PMC90438

- Nordmann
P, Naas T, Poirel L. Global spread of Carbapenemase-producing
Enterobacteriaceae. Emerg Infect Dis. 2011;17:1791–8.
https://doi.org/10.3201/eid1710.110655 PMid:22000347 PMCid:PMC3310682

- Queenan
AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin
Microbiol Rev. 2007;20:440–58. https://doi.org/10.1128/CMR.00001-07
PMid:17630334 PMCid:PMC1932750

- Polat
M, Tapisiz A. Amikacin Monotherapy for Treatment of Febrile Urinary
Tract Infection Caused by Extended-Spectrum β-Lactamase-producing
Escherichia coli in Children. Pediatr Infect Dis J. 2018;37:378-9.
https://doi.org/10.1097/INF.0000000000001860 PMid:29533350

[TOP]


























