Pegdwendé Abel Sorgho1,2, Jeremy James Martinson3, Florencia Wendkuuni Djigma1,2*, Albert Théophane Yonli1,2, Bolni Marius Nagalo4, Tegwinde Rebeca Compaore1,2, Dorcas Obiri-Yeboah5, Birama Diarra1, Herman Karim Sombie1, Arsène Wendpagnangdé Zongo1, Abdoul Karim Ouattara1,2, Serge Théophile R. Soubeiga1,2, Lassina Traore1, Lewis R. Roberts6 and Jacques Simpore1,2.
1 Laboratory
of Molecular Biology and Genetics (LABIOGENE), University Ouaga I Prof.
Joseph Ki-Zerbo, P.O. Box 7021, Ouagadougou 03, Burkina Faso.
2 Pietro Annigoni Biomolecular Research Center (CERBA), P.O. Box 364, Ouagadougou 01, Burkina Faso.
3
Department of Infectious Diseases and Microbiology, University of
Pittsburgh Graduate School of Public Health, 130 De Soto St, Pittsburgh
PA 15261, USA.
4 Division of Hematology & Oncology, Mayo Clinic, Arizona, 13400 E. Shea Blvd. Scottsdale Arizona, 85259, USA.
5 Department of Microbiology and Immunology, School of Medical Sciences, University of Cape Coast, Ghana.
6 Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, Minnesota, 1216 2nd St SW, Rochester, MN 55902, USA.
Correspondence to: Dr. Florencia W. DJIGMA: Department of Biochemistry
and Microbiology, Laboratory of Molecular Biology and Genetic
(LABIOGENE) at University Ouaga I Prof. Joseph Ki-Zerbo, Burkina Faso;
Tel: +226 50-30-70-64/65; Fax: +226 50-30-72-42; Email:
florencia.djigma@gmail.com or
f.djigma@labiogene.org
Published: November 1, 2018
Received: May 25, 2018
Accepted: October 1, 2018
Mediterr J Hematol Infect Dis 2018, 10(1): e2018060 DOI
10.4084/MJHID.2018.060
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background/Objective:
Hepatitis B virus (HBV) infection is the leading risk factor for
cirrhosis and hepatocellular carcinoma (HCC).The objective of this
investigation was to assess the association between "Killer Cell
Immunoglobulin-Like Receptor" (KIR) gene frequencies and chronic HBV
infection.
Methods:
Chronic HBV carriers and healthy patients were selected for this study.
The viral load for HBV were performed, and SSP-PCR was used to
characterize the frequencies of KIR genes.
Results:
The study suggested that inhibitory genes KIR2DL2 (crude OR = 2.82; p ˂
0.001), KIR2DL3 (crude OR = 2.49; p ˂ 0.001) and activator gene KIR2DS2
(crude OR = 3.95; p˂ 0.001) might be associated with chronic stages of
HBV infection. Conversely the inhibitory genes KIR3DL1 (crude OR =
0.49; p = 0.0018) and KIR3DL2 (crude OR = 0.41; p = 0.005), the
activator gene KIR2DS1 (crude OR = 0.48; p = 0.014) and the pseudo gene
KIR2DP1 (crude OR = 0.49; p = 0.008) could be associated with immunity
against HBV infection. Chronic HBV patients who are carriers for the
KIR3DL3 gene (crude OR = 8; p = 0.048) were positive for HBeAg and
patients who carried the KIR3DL2 gene (crude OR = 3.21; p = 0.012) had
a high HBV viral load compared to the rest of the study population.
Conclusion:
Our data showed evidence of a correlation between the risk of
developing chronic HBV infection and certain KIR gene frequencies and
also show that KIR3DL1, KIR3DL2, KIR2DS1 might confer a protective
status against chronic HBV infection.
|
Introduction
Worldwide,
chronic hepatitis B virus (HBV) infection is the leading cause of
cirrhosis and hepatocellular carcinoma (HCC). Several factors might
influence disease progression such as mixed infection or co-infection
with other HBV genotypes or sub-genotypes, hepatitis C (HCV) and host
immunity. To date, there is no accurate method to identify high risk
groups for cirrhosis and HCC in Sub-Saharan Africa. Hepatitis B virus
(HBV) infection is a major life-threatening disease in resource limited
areas where access to vaccination, serological screening, and patient
monitoring are daily challenges. According to the World Health
Organization[1] in 2017, approximately 257 million people are suffering
from chronic HBV infection
(http://www.who.int/mediacentre/factsheets/fs204/en/). Also, every year
roughly 1 million will succumb to chronic HBV (http://www.hepb.org/what-is-hepatitis-b/what-is-hepb/facts-and-figures/).[1]
Chronic hepatitis C virus (HCV) infection is the most common risk
factor for developing HCC in Western countries, but in contrast, both
chronic HBV and hepatitis C (HCV) are highly prevalent in sub-Saharan
Africa, resulting in about a quarter of all HCC cases worldwide.
Sub-Saharan Africa and East Asia have the highest prevalence with about
6.2% of the adult population infected.[1-5] West Africa is a highly
endemic region for HBV infection; the most common route of infection is
a vertical transmission from mother to child followed by sexual
intercourse in the adult population. The limited data on HBV
epidemiology in Burkina Faso displayed a spatial distribution of HBV
prevalence from 9% in Ouagadougou and Bobo-Dioulasso (Central and
Western areas) to 14.4% in Fada N'gourma (Eastern area).[6-10] The
persistence of chronic HBV infection is the main cause for developing
liver cirrhosis and HCC, although, much remains to be learned on the
molecular mechanisms of HBV pathogenesis. The progression of HBV
infection to its chronic stages is associated with a complex interplay
between the virus and its host. In host immunity, viral and epigenetic
factors play a key role in the outcomes of chronic infection,[11,12]
and in some cases, the infected host immune system can manage to
suppress the virus. However, immune evasion strategies allow viral
particles to escape immune clearance, as a consequence of the
evolution of both the immune system and viral epitopes
mutations.[13,14] Natural Killer (NK) cells are cytotoxic lymphocytes,
major components of innate immunity that play an important role in the
immune-mediated rejection process of virally infected cells and tumor
cells.[15] Furthermore, NK cells function by secreting cytokines that
will, in turn, modulate the immune response of the host against viral
infection and aberrant cells by activating the adaptive immune
effectors such as dendritic cells and T lymphocytes.[16] The human KIR
gene locus is located on chromosome 19q13.4 in the Leukocyte Receptor
Complex (LRC) and encodes approximately 15 KIR genes and two pseudo
genes (2DP1, 3DP1).[17,18] These genes are divided in inhibitor genes
(KIR3DL3, KIR2DL2, KIR2DL3, KIR2DL5B, KIR2DL1, KIR3DL1, KIR2DL5A,
KIR3DL2) and activator genes (KIR2DS2, KIR2DS3, KIR3DS1, KIR2DS5A,
KIR2DS5B, KIR2DS4, KIR2DS1); KIR2DL4 gene that can act as either an
activator or inhibitor.[17,19] KIR receptors are glycoproteins found on
the surface of NK cells involved in the activation or inhibition of the
interactions between NK cells and the molecules of the Major
Histocompatibility Complex (MHC) class I.[20,21] Healthy cells
expressing MHC class I proteins are protected through inhibitory
mechanisms that prevent their lysis by “self-recognition”, whereas
cells infected by viruses and cancer cells lacking the MHC class I
molecules on their surfaces are destroyed by lysis activating
receptors.[22] KIR receptors are named according to the number of
extracellular immunoglobulin domains they carry (2D or 3D) and the
length of their cytoplasmic tail which can be long or short (L or
S).The presence of a long cytoplasmic tail with two Immunoreceptor
Tyrosine-based Inhibition Motif (ITIM) confers inhibitory activity to
inhibitory KIRs (2DL, 3DL) and the presence of a short cytoplasmic tail
confers activating activity to KIR activators (2DS, 3DS).[23,24] KIR
genes have been divided into two haplotypes A and B depending on the
presence of specific genes. The latest haplotype definition has
identified that haplotype A is composed of KIR3DL3, KIR2DL3, KIR2DP1,
KIR2DL1, KIR3DP1, KIR2DL4, KIR3DL1, KIR2DS4, and KIR3DL2 genes,
while all other haplotypes as described as haplotype B (14th
International HLA and Immunogenetics Workshop, 2005). Previous studies
have shown the involvement of KIR genes in the pathogenesis of some
diseases such as type 1 diabetes mellitus,[25,26] hepatitis C virus
infection[27,28] and perinatal transmission of HIV infection.[29,30] In
China, a study has shown that the KIR2DS2, KIR2DS3 genes were
associated with chronic hepatitis B virus infection, and the KIR2DS1,
KIR3DS1, KIR2DL5 genes were considered protective genes facilitating
HBV viral clearance.[31] Another report from Turkey showed that KIR2DL3
and KIR3DS1 genes could protect the host against infection with
HBV.[32] Only one study has been conducted in West Africa and
showed that Gambian carriers for the KIR3DS1 gene had a high risk of
being positive for HBeAg as well as carrying a high HBV viral load,
while KIR2DL3 gene carriers had low viral load.[33] In Burkina Faso,
there are no reports on the association between the KIR gene
frequencies and chronic carriage of HBV. Therefore, our pilot study
sought to assess the interplay between the KIR gene frequencies and
chronic HBV infection in the population of Burkina Faso and to identify
which genes are strongly correlated to the risk of developing CHB.
Better management of chronic HBV patient could prevent the rapid onset
of liver cirrhosis and hepatocellular carcinoma in the Burkinabe
population.
Material and Methods
Ethical considerations.
This investigation was approved by the National Health Ethic Committee
of Burkina Faso (reference number No2017-01-004). Patients’ written and
informed consents were obtained according to the Helsinki Declarations.
All results were used as parameters in the therapeutic management of
patients.
Type and population of the study.
This was a prospective study conducted from January to September 2017.
A total of 244 individuals aged 18 and over and divided into two groups
were included in this study. The first group consisted of 110 carriers
for chronic HBV (HBsAg positive > six months) recruited at the
Pietro Annigoni Biomolecular Research Center (CERBA / LABIOGENE). The
second group included 134 negative controls subjects for HBV, HCV, and
HIV recruited at the Regional Blood Transfusion Center of Ouagadougou
(CRTS / O).
Samples collection and measurement of HBV, HCV and HIV viral markers.
Chronic HBV patient blood samples were collected in dry and EDTA tubes
and stored in the Infectious Diseases Research Unit of CERBA. Control
blood samples were collected from healthy volunteer non-remunerated
blood donors at CRTS / O. Serological tests using four-generation ELISA
Ag/Ab were performed for HIV, HCV and HBV screening and confirmation in
the control group using cobas e 411 Analyzer (Roche Diagnostics GmbH
Mannheim Germany) according to the manufacturer's protocol. After
centrifugation at 3,500 rpm for 10 min, plasma was recovered for
determination of HBV viral load and blood pellet for KIR gene research.
The serum of HBV-positive patients was used to screen blood markers of
HBV (HBsAg; HBeAg; anti HBe-Ab); using the HBV One Step Hepatitis B
Virus Combo Test Kit (Abon Biopharm Guangzhou, Co., Ltd. China).
Extraction of viral DNA and Determination of HBV viral load.
Viral DNA was extracted from 200 μL of plasma using the PureLink®
Genomic DNA Extraction Kit (Life Technologies, CA USA) according to the
manufacturer's protocol. The DNA samples were stored at -20°C until
further analysis. Plasma viral load was determined using the 7500Fast
Real Time PCR system (Applied Biosystems, USA) using the Genesig HBV
Real Time Quantitative Kit Primer design kit (Southampton, United
Kingdom).
Genomic DNA Extraction and Determination of KIR Genes by SSP-PCR (Sequences Specific Primer).
Genomic DNA was extracted from the whole blood using the salting-out
method and stored at -80°C until analysis as previously described.[34]
DNA purity and concentration were determined using a Biodrop (Isogen
Life Science, NV/S.A, Temse, Belgium). Approximatively 100 ng/μl
of DNA was used to amplify the subset of 12 targeted KIR genes using
the SSP-PCR method as previously described.[35] The PCR reactions were
performed in 60 µL of the reaction mixture containing 100 ng/µL of DNA
(variable volume), 7.5 μL of 10 × CPR buffer, 2.25 μL MgCl2; 0.6 μL of
dNTPs and 0.375 μL of PlatinumTM DNA Taq polymerase in nuclease-free
water.[35]
The PCR reactions were performed as follows: after initial
denaturation for 3 min at 94°C, the amplifications were carried out
respectively for 5 cycles, 21 cycles and 4 cycles of denaturation at
94°C, annealing at primer specific temperature for 15 sec (65°C and
60°C) or 1 min (55°C for 4 cycles step), and extension at 30 sec at
72°C or 2 min for 4 cycles step with a final extension at 72°C for 7
min. The PCR products were separated on 3% agarose gel and visualized
under UV light at 312 nm using the Gene flash apparatus (Gene Flash
syngenge Bio Imaging, USA). PCR products were validated against a
positive internal control corresponding to the DRB1 gene
fragment.
Statistical analysis.
Standard Statistical Package for Social Sciences (SPSS) version 20.0
was used for data analysis and interpretation. Changes were considered
statistically significant at p ≤ 0.05, using the Fisher Exact test.
Odds ratio (OR) and confidence intervals (CI) at 95% were calculated to
estimate the associations between the KIR gene frequencies and HBV
chronic infection using Epi Info 7.
Results
Sociodemographic characteristics of the study population.
The study population comprises a total of 244 people (110 chronic HBV
patients and 134 control subjects) aged from 18 to 73, with an average
age of 34.75 ± 11.57. The mean age of the HBV patients was 38.55 ±
13.07 years and 31.62 ± 9.11 years in the control group. The percentage
of men was 48.77% (119/244) and 51.23% (125/244) for women with a sex
ratio of 0.95. The most heavily represented age groups in the study
population were 25 to 39 years old with 51.23% (125/244) and the
difference was statically significant (p = 0.017). There were slightly
more men [53.64% (59/110)] infected with chronic HBV than women [46.36%
(51/110)], as shown in table 1.
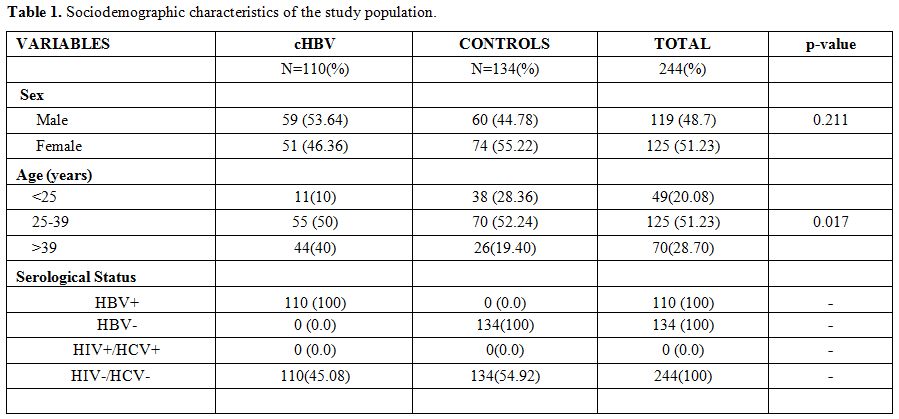 |
Table 1. Sociodemographic characteristics of the study population. |
Biochemical and virologic features of chronic HBV patients.
Liver function was assessed using the measurement of Alanine
aminotransferase (ALT) and aspartate aminotransferase (AST) enzyme
levels, and hepatitis B infection was determined by measurements of
viral envelope antigen (HBeAg) and antibody (anti-HBeAb) levels. We
tested HBsAg in all HBV chronic patients to confirm chronic HBV
infection, and HIV, HBV, HCV tests were performed in controlled
subjects to rule out possible cases of infection with these viruses.
Viral load was below 2000 IU/ml for 57.3%, (63/110) of patients, and
greater than 2000 IU/ml in 42.7% (47/110) of patients (Table 2).
17.27%, (19/110) of the patients were positive for HBeAg and negative
for anti-HBeAb, suggestive of replicating virus, while 82.73% (91/110)
of patients were negative for HBeAg and positive for anti-HBeAg,
indicating chronic infection. In chronic hepatitis B patients who had
HBeAg positive, 94.7% or (18/19) had a viral load greater than or equal
to 2000 IU/mL compared to 5.3% (1/19) who had a viral load less than
2000 IU/mL. The univariate analysis showed that HBeAg is strongly
associated with an increased viral load (Crude OR = 38.48, p <
0.001) and a multivariate age-sex-matched analysis supports that HBeAg
is associated with increased HBV viral load (Adj -OR = 31.1, p =
0.002). A total of 34 patients had alanine aminotransferase (ALT)
levels greater than 40 IU/mL, and 76 patients had ALT levels less than
or equal to 40 IU/mL. Among patients who had ALT levels greater than 40
IU/mL, 82.4% (28/34) had a viral load greater than or equal to 2000
UI/mL versus 17.6% (6/34) who had a viral load less than 2000 IU/mL.
Univariate analysis shows that elevated ALT levels are associated with
increased HBV viral load (crude OR = 14, p < 0.001), a multivariate
age-sex-matched analysis supports the assumption that elevation of ALT
levels is associated with high values of HBV viral load (Adj -OR =
12.02, p < 0.001).
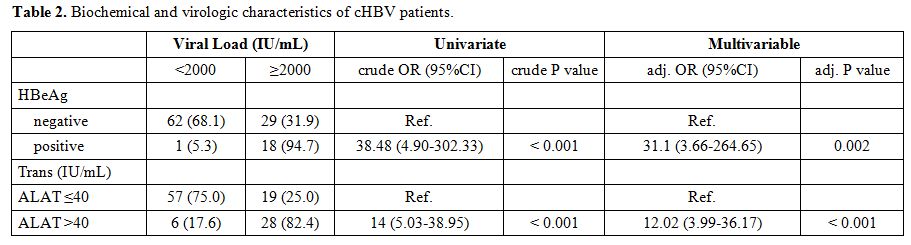 |
Table
2. Biochemical and virologic characteristics of cHBV patients. |
Characterization of KIR genes by SSP-PCR.
A total of 16 KIR genes were determined by PCR-SSP. Our results shown
the different frequencies of KIR genes between chronic HBV patients and
controls subjects in this study (Table 3).
Thus a univariate analysis shown that KIR genes such as KIR2DL2 (crude
OR = 2.82 , 95%CI =1.61-4.96 , p < 0.001), KIR2DL3 (crude OR = 2.49
, 95%CI = 1.47-4.21 , p ˂ 0.001) and KIR2DS2 (crude OR = 3.95 , 95%CI =
2.15-7.27, p < 0.001) were more frequent in chronic hepatitis
B carriers than in control subjects however the KIR genes such as
KIR3DL1 (crude OR = 0.49 , 95%CI = 0.28-0.89, p = 0.0018) and KIR3DL2
(crude OR = 0.41, 95%CI = 0.22-0.77, p = 0.005); KIR2DS1 (crude OR =
0.48, 95%CI = 0.27-0.86, p = 0.014) and the pseudo gene KIR2DP1 (crude
OR = 0.49, 95%CI = 0.29-0.83, p = 0.008) were more frequent in controls
than in chronic hepatitis B carriers (Table 3).
An association has been found between the KIR genes and the viral
replication marker of HBV, the univariate analysis shown that the
carriers of the KIR3DL3 gene are likely to be HBeAg positive (crude OR
= 8, 95%CI = 1.02-62.91, p = 0.048) (Table 4).
We found a high frequency of the KIR3DL2 gene in chronic HBV patients
who had a viral load greater than or equal to 2000 IU/mL, the
univariate analysis shows that the KIR3DL2 gene is associated with the
increase in the viral load HBV (crude OR = 3.21, 95% CI = 1.29-7.99, p
= 0.012) (Table 5).
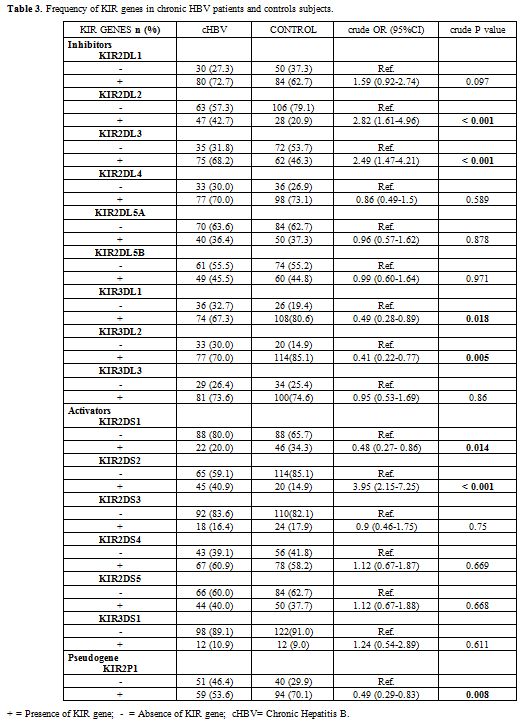 |
Table 3.
Frequency of KIR genes in chronic HBV patients and controls subjects. |
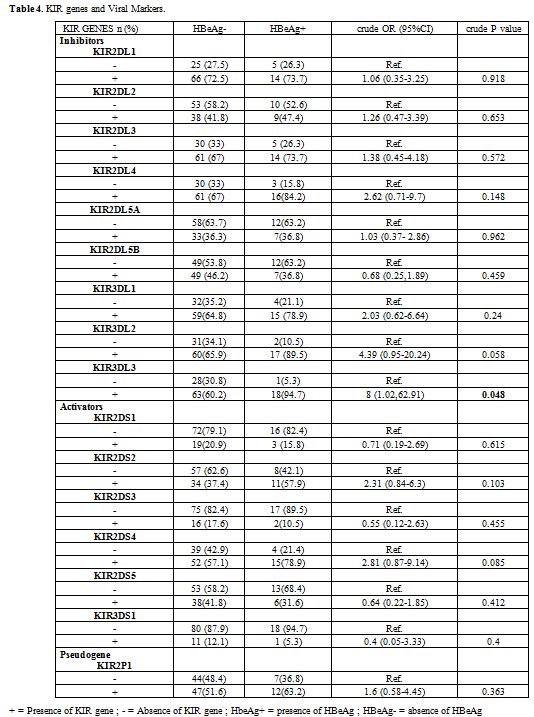 |
Table
4. KIR genes and Viral Markers. |
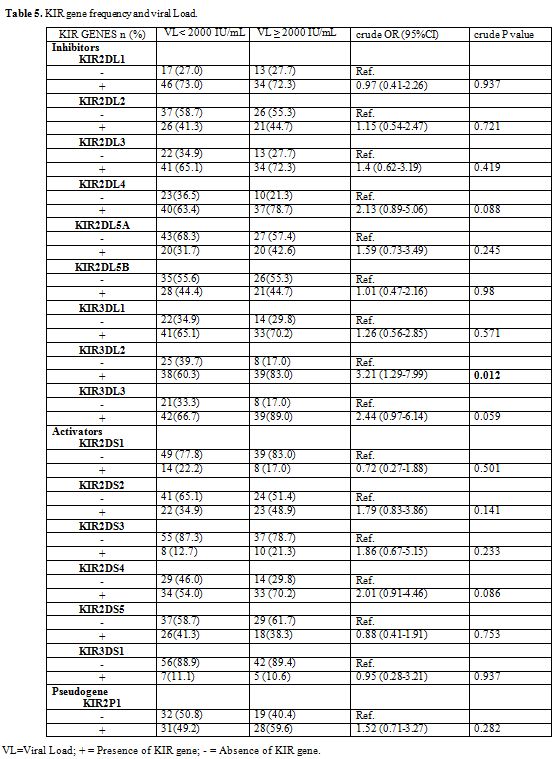 |
Table 5. KIR gene frequency and viral Load. |
Discussion
Our
study consisted in characterizing for the first time in a population of
Burkina Faso, the KIR genes. The choice of the chronic hepatitis B
population and negative blood donors for the HIV, HCV and HBV test
allowed us to have a fairly representative sample of the general
population of Burkina Faso, a country endemic to Hepatitis B1 with a
prevalence of almost 15%.[6-9,36,37] In addition, this choice is in the
wake of studies that seek to understand how genetic factors and cells
of the innate immune system are involved in cases of virus
infection.[38-40] In Burkina Faso, a significant proportion of patients
with chronic hepatitis B (cHBV) acquired the infection early in life
through mother-to-child transmission.[41,42] The average age of our
study population was 34.75±11.57 years. The most represented age group
in our study population was 25 to 39 (51.23%) (p=0.017). In this group,
they were a large number of women who were diagnosed following an
antenatal check-up.
We have found that the presence of the viral
replication marker is associated with viral loads greater than or equal
to 2000 IU/mL (crude OR=38.48; p˂0.001). As in others studies that have
demonstrated that HBeAg detection is associated with hepatitis B virus
replication;[43,44] a multivariate analysis adjusted for age and sex
shows an association between HBeAg and the viral load greater than or
equal to 2000 IU/mL (Adj-OR=31.1 p=0.002). Similarly, alanine
aminotransferase (ALT) levels greater than or equal to 40 IU/mL in
chronic HBV patients were associated with viral load values greater
than or equal to 2000 IU/mL (crude OR=14; p˂ 0,001), adjusted for age
and sex we found a significant association (Adj-OR=12.02 p˂0.001).
The
main limitation of our study is that we have characterized only KIR
genes, but not the KIR/HLA combination. The frequency of inhibitory
genes KIR2DL2, KIR2DL3, and activator gene KIR2DS2 was high in chronic
HBV patients than in control group while the frequency of inhibitory
genes KIR3DL1, KIR3DL2, the activator gene KIR2DS1, and the pseudo gene
KIR2DP1 were high in the control group than in chronic HBV patients. In
our study, the KIR genes, KIR2DL2, KIR2DL3, KIR2DS2, are associated
with HBV chronic infection, and KIR3DL1, KIR3DL2, KIR2DS2, KIR2P1 are
associated with protection against chronic HBV infection. The KIR3DL3
gene was linked to the HBeAg positive status of patients. There was a
statistically significant correlation between the presence of KIR3DL2
gene and a high viral load of HBV. A Chinese study suggested that the
activating genes KIR2DS2 and KIR2DS3 genes could be associated with
chronic HBV infection, which induced a persistent yet weak inflammatory
reaction that results in continuous injury of live tissues on chronic
hepatitis and that inhibitory genes KIR2DL5 and activators genes
KIR2DS1, KIR3DS1 could protect, thus facilitate HBV viral
immune clearance.[31] On the other hand, a Turkish study showed that
the inhibitory gene KIR2DL3 and activator gene KIR3DS1 could be
protective against HBV infection.[32] Alongside this Turkish study; Gao
et al. (2010) found that the combination of KIR2DL3 and HLA-C1
conferred protection against HBV infection and that the combination
KIR2DL1 and HLA-C2 could be associated with HBV infection;[45] Di Bona
et al. (2017) found that KIR ligand group HLA-A-Bw4 and HLA-C2 are
associated with HBV chronic infection. Subjects possessing these
alleles are more susceptible to be HBV chronic carriers while KIR2DL3
confer protection against HBV chronic infection.[46]
In our
study, patients who carried the KIR3DL3 gene were associated with HBeAg
positive status which is a viral replication marker of HBV. Whereas,
those who carried the inhibitory KIR3DL2 gene had a high HBV viral
load. In the Gambia, a study showed that carriers of the KIR3DS1 gene
were HBV-positive and had a high viral load while KIR2DL3 gene carriers
had a low HBV viral load.[33] Genotypes and haplotypes containing more
activator genes would play an essential role in chronic infection or
elimination of HBV.[47] Combinations of KIR genes and HLA molecules are
associated with the development of hepatocellular carcinoma in patients
infected with chronic hepatitis B virus.[48]
Conclusion
This
investigation showed that KIR inhibitory genes KIR2DL2, KIR2DL3, and
KIR2DS2 activator are associated with chronic HBV infection, while the
inhibitory genes KIR3DL1, KIR3DL2, the activator gene KIR2DS1, and the
pseudo gene KIR2DP1 are associated with protection against chronic
infection by HBV. Also, the KIR3DL3 gene was linked to the HBeAg
positive status; while the KIR3DL2 gene was associated with the
evolution of HBV viral load in the context of Burkina Faso. However,
KIR/HLA studies combined with additional genotyping of HBV are needed
to investigate the molecular mechanisms by which KIR genes contribute
to the infection or elimination of the hepatitis B virus.
Aknowledgement
We
would like to thank the Institute of International Education, the U.S.
Department of State's Bureau of Educational and Cultural Affairs, The
US embassy of Ouagadougou and the Council for International Exchange of
Scholars (CIES) for funding this project through the J. William
Fulbright Foreign Scholarship (African Research Scholar Program).
References
- OMS, Aide mémoire N° 204, Juillet 2017.
http://www.who.int/mediacentre/factsheets/fs204/fr/index.html
(published 30/01/2017).
- Zampino R, Boemio A, Sagnelli C, Alessio
L, Adinolfi LE, Sagnelli E, Coppola N. Hepatitis B virus burden in
developing countries. World J Gastroenterol. 2015; 21 (42): 11941-53.
https://doi.org/10.3748/wjg.v21.i42.11941 PMid:26576083
PMCid:PMC4641116
- Stasi C, Silvestri C, Voller F. Emerging Trends
in Epidemiology of Hepatitis B Virus Infection. J Clin Transl Hepatol,
2017; 5(3): 272-276. https://doi.org/10.1016/j.jceh.2015.06.002
- Petruzziello
A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV)
Related Hepatocellular Carcinoma. Open Virol J. 2018; 12: 26-32.
https://doi.org/10.2174/1874357901812010026 PMid:29541276
PMCid:PMC5842386
- Lemoine M, Nayagam S, Thursz M. Viral hepatitis
in resource-limited countries and access to antiviral therapies:
current and future challenges. Future Virol. 2013; 8(4): 371-380.
https://doi.org/10.2217/fvl.13.11 PMid:23662157 PMCid:PMC3646239
- Tao
I, Compaoré TR, Diarra B, Djigma F, Zohoncon TM, Assih M, Ouermi D,
Pietra V, Karou SD, Simpore J. Seroepidemiology of hepatitis B and C
viruses in the general population of burkina faso. Hepat Res Treat.
2014; 781843. Epub 2014 Aug 5. https://doi.org/10.1155/2014/781843
- Sanou AM, Benkirane K, Tinto B, Cissé A, Sagna T, Ilboudo
AK, Dording C, Tarnagda Z, Muller CP, Hübschen JM. Prevalence of
Hepatitis B virus and Hepatitis D virus coinfection in Western Burkina
Faso and molecular characterization of the detected virus strains. Int
J Infect Dis. 2018; 70:15-19 https://doi.org/10.1016/j.ijid.2018.02.004
PMid:29432880
- Nagalo MB, Sanou M, Bisseye C, Kaboré MI, Nebie
YK, Kienou K, Kiba A, Dahourou H, Ouattara S, Zongo JD, Simporé J.
Seroprevalence of a human immunodeficiency virus, hepatitis B and C
viruses and syphilis among blood donors in Koudougou (Burkina Faso) in
2009. Blood Transfus. 2011; 9(4): 419-24. PMid:21839011
PMCid:PMC3200412
- Nagalo BM, Bisseye C, Sanou M, Kienou K, Nebié
YK, Kiba A, Dahourou H, Ouattara S, Nikiema JB, Moret R, Zongo JD,
Simpore J. Seroprevalence and incidence of transfusion-transmitted
infectious diseases among blood donors from regional blood transfusion
centres in Burkina Faso, West Africa. Trop Med Int Health. 2012; 17(2):
247-53. https://doi.org/10.1111/j.1365-3156.2011.02902.x
PMid:21988100
- Ilboudo D, Simpore J, Ouermi D, Bisseye C, Sagna
T, Odolini S, Buelli F, Pietra V, Pignatelli S, Gnoula C, Nikiema JB,
Musumeci S. Towards the complete eradication of mother-to-child HIV/HBV
coinfection at Saint Camille Medical Centre in Burkina Faso, Africa.
Braz J Infect Dis. 2010; 14(3): 219-24.
https://doi.org/10.1016/S1413-8670(10)70047-7
- Rongrui L, Na
H, Zongfang L, Fanpu J, Shiwen J. Epigenetic mechanism involved in the
HBV/HCV-related hepatocellular carcinoma tumorigenesis. Curr Pharm Des.
2014; 20(11): 1715-25. https://doi.org/10.2174/13816128113199990533
PMid:23888939
- Hong X., Kim ES, Guo H. Epigenetic regulation of
hepatitis B virus covalently closed circular DNA: Implications for
epigenetic therapy against chronic hepatitis B. Hepatology. 2017;
66(6): 2066-2077. https://doi.org/10.1002/hep.29479 PMid:28833361
- Lok
AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009;
50(3): 661-2. https://doi.org/10.1002/hep.23190 PMid:19714720
- Zhang
Y, Wu Y, Deng M, Xu D, Li X, Xu Z, Hu J, Zhang H, Liu K, Zhao Y, Gao F,
Bi S, Gao GF, Zhao J, Liu WJ, Meng S. CD8(+) T-Cell Response-Associated
Evolution of Hepatitis B Virus Core Protein and Disease Progress. J
Virol. 2018 Aug 16;92(17). pii: e02120-17. Print 2018 Sep 1
https://doi.org/10.1128/JVI.02120-17
- Hamerman JA, Ogasawara
K, Lanier LL. NK cells in innate immunity. Curr Opin Immunol. 2005;
17(1): 29-35. https://doi.org/10.1016/j.coi.2004.11.001 PMid:15653307
- White-Grindley E, Si K. RISC-y Memories. Cell. 2006; 124(1): 23-6. https://doi.org/10.1016/j.cell.2005.12.027 PMid:16413478
- González-Galarza
FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, Da Silva AL, Teles e
Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, Middleton D. Allele
frequency net 2015 update: new features for HLA epitopes, KIR and
disease and HLA adverse drug reaction associations. Nucleic Acids Res.
2015; 43 (Database issue): p. D784-8 Epub 2014 Nov 20.
- Takeshita
LY, Gonzalez-Galarza FF, dos Santos EJ, Maia MH, Rahman MM, Zain SM,
Middleton D, Jones AR. A database for curating the associations between
killer cell immunoglobulin-like receptors and diseases in worldwide
populations. Database (Oxford), 2013. bat021..
https://doi.org/10.1093/database/bat021
- Faure M, Long EO.
KIR2DL4 (CD158d), an NK cell-activating receptor with inhibitory
potential. J Immunol. 2002; 168(12): 6208-14.
https://doi.org/10.4049/jimmunol.168.12.6208 PMid:12055234
- Vilches
C, Parham P. KIR: diverse, rapidly evolving receptors of innate and
adaptive immunity. Annu Rev Immunol. 2002; 20: 217-51.
https://doi.org/10.1146/annurev.immunol.20.092501.134942
PMid:11861603
- Middleton D, Williams F, Halfpenny IA. KIR genes.
Transpl Immunol. 2005; 14(3-4): 135-42.
https://doi.org/10.1016/j.trim.2005.03.002 PMid:15982555
- Jonsson
AH, Yokoyama WM. Natural killer cell tolerance licensing and other
mechanisms. Adv Immunol. 2009; 101: 27-79.
https://doi.org/10.1016/S0065-2776(08)01002-X
- Selvakumar A,
Steffens U, Dupont B. Polymorphism and domain variability of human
killer cell inhibitory receptors. Immunol Rev. 1997; 155: 183-96.
https://doi.org/10.1111/j.1600-065X.1997.tb00951.x PMid:9059894
- Moretta
A, Sivori S, Vitale M, Pende D, Morelli L, Augugliaro R, Bottino C,
Moretta L. Existence of both inhibitory (p58) and activatory (p50)
receptors for HLA-C molecules in human natural killer cells. J Exp Med.
1995; 182(3): 875-84. https://doi.org/10.1084/jem.182.3.875
PMid:7650491
- Van der Slik AR, Koeleman BP, Verduijn W, Bruining
GJ, Roep BO, Giphart MJ. KIR in type 1 diabetes: disparate distribution
of activating and inhibitory natural killer cell receptors in patients
versus HLA-matched control subjects. Diabetes. 2003; 52(10): 2639-42.
https://doi.org/10.2337/diabetes.52.10.2639 PMid:14514651
- Van
der Slik AR, Alizadeh BZ, Koeleman BP, Roep BO, Giphart MJ. Modelling
KIR-HLA genotype disparities in type 1 diabetes. Tissue Antigens. 2007;
69 Suppl 1:101-5.
https://doi.org/10.1111/j.1399-0039.2006.762_5.x PMid:17445178
- Shan
Z, Huang J, Liao Q, Huang K, Wang M, Xu R, Tang X, Zhang W, Nelson K,
Fu Y, Li C, Rong X. Association of killer cell immunoglobulin-like
receptors with spontaneous clearance of hepatitis C virus in the
Chinese population. Transfusion. 2018; 58(4):1028-1035
https://doi.org/10.1111/trf.14527 PMid:29446443
- Khakoo SI,
Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert
JJ, Vlahov D, Hilgartner M, Cox S, Little AM, Alexander GJ, Cramp ME,
O'Brien SJ, Rosenberg WM, Thomas DL, Carrington M. HLA and NK cell
inhibitory receptor genes in resolving hepatitis C virus infection.
Science. 2004; 305(5685): 872-4.
https://doi.org/10.1126/science.1097670 PMid:15297676
- Zwolińska
K, Błachowicz O, Tomczyk T, Knysz B, Gąsiorowski J, Zalewska M,
Orzechowska BU, Sochocka M, Piasecki E. The effects of killer cell
immunoglobulin-like receptor (KIR) genes on susceptibility to HIV-1
infection in the Polish population. Immunogenetics. 2016; 68(5):
327-37. https://doi.org/10.1007/s00251-016-0906-1 PMid:26888639
PMCid:PMC4842214
- Omosun YO, Blackstock AJ, Williamson J, Van
Eijk AM, Ayisi J, Otieno J, Lal RB, Ter Kuile FO, Slutsker L, Shi YP.
Association of maternal KIR gene content polymorphisms with reduction
in perinatal transmission of HIV-1. PLoS One. 2018; 13(1):
e0191733.Epub 2018 Jan 23.
- Zhi-ming L, Yu-lian J, Zhao-lei F,
Chun-xiao W, Zhen-fang D, Bing-chang Z, Yue-ran Z. Polymorphisms of
killer cell immunoglobulin-like receptor gene: possible association
with susceptibility to or clearance of hepatitis B virus infection in
Chinese Han population. Croat Med J. 2007; 48(6): 800-6.
https://doi.org/10.3325/cmj.2007.6.800 PMid:18074414 PMCid:PMC2213808
- Kibar
F, Goruroglu Ozturk O, Ulu A, Erken E, Inal S, Dinkci S, Kurtaran B,
Tasova Y, Aksu HS, Yaman A. Role of KIR genes and genotypes in
susceptibility to or protection against hepatitis B virus infection in
a Turkish cohort. Med Sci Monit. 2014; 20: 28-34.
https://doi.org/10.12659/MSM.889893 PMid:24407110 PMCid:PMC3894916
- Yindom
LM, Mendy M, Bodimeade C, Chambion C, Aka P, Whittle HC, Rowland-Jones
SL, Walton R. KIR content genotypes associate with carriage of
hepatitis B surface antigen, e antigen and HBV viral load in Gambians.
PLoS One. 2017; 12(11): e0188307. 2017 Epub Nov 17.
- Miller SA,
Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA
from human nucleated cells. Nucleic Acids Res. 1988; 16(3): 1215.
https://doi.org/10.1093/nar/16.3.1215 PMid:3344216
PMCid:PMC334765
- Kulkarni S, Martin MP, Carrington M. KIR
genotyping by multiplex PCR-SSP. Methods Mol Biol. 2010; 612: 365-75.
https://doi.org/10.1007/978-1-60761-362-6_25 PMid:20033654
PMCid:PMC3464904
- Diarra B, Yonli AT, Sorgho PA, Compaore TR,
Ouattara AK, Zongo WA, Tao I, Traore L1, Soubeiga ST, Djigma FW,
Obiri-Yeboah D, Nagalo BM, Pietra V, Sanogo R, Simpore J. Occult
Hepatitis B Virus Infection and Associated Genotypes among
HBsAg-negative Subjects in Burkina Faso. Mediterr J Hematol Infect Dis,
2018. 10(1): p. e2018007.
- Simpore J, Savadogo A, Ilboudo D,
Nadambega MC, Esposito M, Yara J, Pignatelli S, Pietra V, Musumeci S.
Toxoplasma gondii, HCV, and HBV seroprevalence and co-infection among
HIV-positive and -negative pregnant women in Burkina Faso. J Med Virol
2006. 78(6): 730-733. https://doi.org/10.1002/jmv.20615 PMid:16628587
- Compaore
TR, Soubeiga ST, Ouattara AK, Obiri-Yeboah D, Tchelougou D, Maiga M,
Assih M, Bisseye C, Bakouan D, Compaore IP, Dembele A, Martinson J,
Simpore J., APOBEC3G Variants and Protection against HIV-1 Infection in
Burkina Faso. PLoS One, 2016. 11(1): p. e0146386.
https://doi.org/10.1371/journal.pone.0146386 PMid:26741797
PMCid:PMC4704832
- Compaore TR, Diarra B, Assih M, Obiri-Yeboah D,
Soubeiga ST, Ouattara AK, Tchelougou D, Bisseye C, Bakouan DR, Compaore
IP, Dembele A, Djigma WF, Simpore J. HBV/HIV co-infection and APOBEC3G
polymorphisms in a population from Burkina Faso. BMC Infect Dis, 2016.
16: p. 336. https://doi.org/10.1186/s12879-016-1672-2 PMid:27449138
PMCid:PMC4957463
- Laaribi AB, Hannachi N, Ben Yahia H, Marzouk M,
Mehri A, Belhadj M, Yacoub S, Letaief A, Ouzari HI, Boudabous A,
Boukadida J, Rizzo R, Zidi I. Human leukocyte antigen (HLA-F)
polymorphism is associated with chronic HBV infection. 3 Biotech, 2018.
8(1): p. 49
- Sangaré L, Sombié R, Combasséré AW, Kouanda A, Kania
D, Zerbo O, Lankoandé J. [Antenatal transmission of hepatitis B virus
in an area of HIV moderate prevalence, Burkina Faso]. Bull Soc Pathol
Exot, 2009. 102(4): p. 226-9. PMid:19950539
- Ilboudo D., A.
Sawadogo, and J. Simpore. Mother-to-child transmission of hepatitis B
virus, in Ouagadougou, Burkina Faso. Med Trop, 2002. 62(1): p. 99-101
- Hudu
SA, Niazlin MT, Nordin SA, Saeed MI, Tan SS, Omar H, Shahar H, Sekawi
Z. Quantitative Hepatitis B e Antigen: A Better Predictor of Hepatitis
B Virus DNA than Quantitative Hepatitis B Surface Antigen. Clin Lab,
2018. 64(4): p. 443-449. https://doi.org/10.7754/Clin.Lab.2017.170916
- Thompson AJV, Nguyen T, Iser D, Ayres A, Jackson K,
Littlejohn M, Slavin J, Bowden S, Gane EJ, Abbott W, Lau GKK, Lewin S,
Visvanathan K, Desmond PV, Locarnini SA. Serum hepatitis B surface
antigen and hepatitis B e antigen titers: disease phase influences
correlation with viral load and intrahepatic hepatitis B virus markers.
Hepatology, 2010. 51(6): p. 1933-44.
https://doi.org/10.1002/hep.23571 PMid:20512987
- Gao X,
Jiao Y, Wang L, Liu X, Sun W, Cui B, Chen Z, Zhao Y. Inhibitory KIR and
specific HLA-C gene combinations confer susceptibility to or protection
against chronic hepatitis B. Clin Immunol, 2010. 137(1): p. 139-46
https://doi.org/10.1016/j.clim.2010.05.011 PMid:20643584
- Di Bona
D, Aiello A, Colomba C, Bilancia M, Accardi G, Rubino R, Giannitrapani
L, Tuttolomondo A, Cascio A, Caiaffa MF, Rizzo S, Di Lorenzo G, Candore
G, Duro G, Macchia L, Montalto G, Caruso C, KIRIIND (KIR Infectious and
Inflammatory Diseases) Collaborative Group. KIR2DL3 and the KIR ligand
groups HLA-A-Bw4 and HLA-C2 predict the outcome of hepatitis B virus
infection. J Viral Hepat, 2017. 24(9): p. 768-775.
https://doi.org/10.1111/jvh.12698 PMid:28211154
- Lu Z, Zhang B,
Chen S, Gai Z, Feng Z, Liu X, Liu Y, Wen X, Li L, Jiao Y, Ma C, Shao S,
Cui X, Chen G, Li J, Zhao Y. Association of KIR genotypes and
haplotypes with susceptibility to chronic hepatitis B virus infection
in Chinese Han population. Cell Mol Immunol, 2008. 5(6): 457-63.
https://doi.org/10.1038/cmi.2008.57 PMid:19118512
PMCid:PMC4072426
- Pan N, Jiang W, Sun H, Miao F, Qiu
J, Jin H, Xu J, Shi Q, Xie W, Zhang J. KIR and HLA loci are associated
with hepatocellular carcinoma development in patients with hepatitis B
virus infection: a case-control study. PLoS One, 2011. 6(10): p.
e25682. https://doi.org/10.1371/journal.pone.0025682 PMid:21998681
PMCid:PMC3187788
[TOP]




