Nishit Gupta#1, Ravikiran Pawar#1, Sambhunath Banerjee1, Subhajit Brahma1, Asish Rath1, Sundar Shewale1, Mayur Parihar2, Manish Singh2, Arun S R2, Shekhar Krishnan3, Arpita Bhatacharyya3, Anirban Das3, Jeevan Kumar4, Saurabh Bhave4, Vivek Radhakrishnan4, Reena Nair4, Mammen Chandy4, Neeraj Arora5 and Deepak Mishra5.
# Equal Contribution
1 Department of Laboratory Hematology
2 Department of Laboratory Hematology and Cytogenetics.
3 Department of Pediatric Oncology
4 Department of Clinical Hematology
5 Department of Laboratory Hematology and Molecular Genetics, Tata Medical Center, Kolkata
Correspondence to: Dr. Deepak Mishra. Department of Laboratory
Hematology and Molecular genetics, Tata Medical Center,
Kolkata, 14 MAR (E-W),
Rajarhat, West Bengal, India,
700160. Tel: +91 9831132365, +91
33 66057754. E-mail:
deepak.mishra@tmckolkata.com
Published: March 1, 2019
Received: September 17, 2018
Accepted: January 19, 2019
Mediterr J Hematol Infect Dis 2019, 11(1): e2019017 DOI
10.4084/MJHID.2019.017
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background: For diagnosis, sub-categorization and follow up of Acute Leukemia (AL), phenotypic analysis using flow cytometry is mandatory.
Material and methods:
We retrospectively analyzed immunophenotypic data along with
cytogenetics/molecular genetics data (wherever available) from 631
consecutive cases of AL diagnosed at our flow cytometry laboratory from
January 2014 to August 2017.
Results:
Of the total 631 cases, 52.9% (n=334) were acute lymphoblastic leukemia
(ALL), 43.9% (n=277) acute myeloid leukemia (AML), 2.2% (n=14) mixed
phenotypic acute leukemia (MPAL), 0.5% (n=3) acute undifferentiated
leukemia (AUL) and 0.5% (n=3) chronic myeloid leukemia in blast crisis
(CML-BC). ALL cases comprised of 81.7% (n=273/334) B-cell ALLs (95.2%,
n=260/273 common B-ALLs and 4.8%, n=13/273 Pro B-ALLs). CD13 was the
commonest cross lineage antigen, expressed in B-ALL (25.6%, n=70/273),
followed by CD33 (17.9%, n=49) and combined CD13/CD33 (11.3%, n=31/273)
expression. T-ALLs constituted 18.3% (n=61/334) of total ALLs and
included 27.9% (n=17/61) cortical T- ALLs. CD13 was commonest (32.7%,
n=20/61) aberrantly expressed antigen in T-ALLs, followed by CD117
(19.1%, n=9/47). AML cases included 32.1% (n=89/277) AML with recurrent
genetic abnormalities, 9.0% (n=25/277) with FLT3/NPM1c mutation and
58.9% (n=163/277) AML NOS including 14.7% (n=24/163) AML M4/M5, 1.8%
(n=3/163) AML M6 and 3.7% (n=6/163) AML M7. In AMLs, CD19 aberrancy was
the most common (20.2%, n=56/277) followed by CD56 (15.8%, n=42/265).
Conclusions:
In this study, we document the spectrum, correlate the immunophenotype
with genetic data of all leukemias, especially concerning T-ALL where
the data from India is scarce.
|
Introduction
Acute
leukemia (AL) is a clonal hematopoietic stem cell disorder
characterized by an increase in immature cells (≥20%) in peripheral
blood and/or bone marrow. Many classification systems have been
proposed for AL. The first internationally accepted classification was
the French–American–British (FAB) cooperative Group Classification
(1976), entirely based on morphologic criteria. It was subsequently
refined in 1981 and 1985.[1] These modifications of
the classification system did not incorporate the characteristic
immunophenotypic features seen in AL. In 2008, WHO gave a comprehensive
classification system of AL (including morphology, cytochemistry,
immunophenotyping, fluorescence in-situ hybridization (FISH), and
reverse transcriptase polymerase chain reaction (RT-PCR).[2]
Flow
cytometric immunophenotyping is the backbone of WHO classification and
plays the most crucial role in the diagnosis, lineage characterization
and sub-classification of AL. It also provides prognostic as well as
predictive information aiding in modulating therapy appropriately.
Leukemic blasts with specific genetic signatures show peculiar
immunophenotype on flow cytometry. Knowledge and recognition of these
associations can help save time and money, particularly in
resource-constrained settings.[2,3]
In this
study, we retrospectively analyzed 631 consecutive cases of AL
diagnosed at our center, focusing on the spectrum of immunophenotypic
features, aberrancy profiles and their correlation with cytogenetic/
molecular findings wherever available. Published data on
immunophenotypic patterns associated with certain newer entities like
ETP-ALL (Early T-cell precursor lymphoblastic leukemia) along with the
importance of CD1a in T-ALL lack in the literature, especially from the
Indian subcontinent. We have tried to incorporate the immunophenotypic
signatures associated with these entities as well.
Material and methods
This
study is a retrospective analysis of 631 consecutive cases of AL
diagnosed by flow cytometry laboratory from January 2014 to August
2017. The diagnosis was given as per the WHO 2008 criteria.[2]
Peripheral blood smears and bone marrow aspirates were air dried and
stained with Wright-Giemsa stain. Myeloperoxidase was done routinely in
all cases. Other cytochemical stains like periodic acid-Schiff (PAS);
non-specific esterase and specific esterase were performed as and when
required based on the morphological details of the leukemic blasts.[4] Immunophenotyping data was correlated with cytogenetics / molecular genetics data wherever available.
Flow cytometry.
Immunophenotyping was performed either on bone marrow (n=504) or
peripheral blood (n=127) samples received in EDTA. The six-color
analysis was performed on FACSCanto II (Becton Dickinson, San Jose, CA,
USA) using the following fluorochromes; FITC (Fluorescein
isothiocyanate), PE (Phycoerythrin), PerCP Cy5.5, APC
(Allophycocyanin), PE-Cy7 and APC-H7. Two to four ml of EDTA sample was
taken for the immunophenotypic analysis depending on the sample cell
count. The standard technique of antibody staining (incubation at room
temperature, 15-20 minutes) followed by lysis and washing was used.
Cytoplasmic markers were permeabilized with BD FACS™ Permeabilizing
Solution 2 (Becton Dickenson, San Jose, CA, USA). For data analysis and
interpretation (BD FACS Diva version 6.1.3 software), a cut off of 20%
expression for surface marker positivity and 10% for cytoplasmic
markers was used as per the EGIL criteria.[2] For all
AL cases, three tubes were used in common: 1) CD7 FITC, CD117 PE, CD34
PerCP Cy5.5, CD19 PE-Cy7, CD10 APC, CD45 APC-H7; 2) CD5 FITC, CD13 PE,
CD20 PerCP Cy5.5, HLA-DR PE-Cy7, CD33 APC, CD45 APC-H7; 3) cMPO FITC,
cCD79a PE, cCD3 PE-Cy7, TdT APC, CD45 APC-H7. Characterization of
blasts was done based on CD45/ side scatter along with other markers
like CD34, CD117, CD10, and CD19. Once delineated into either AML,
B-ALL or T-ALL based on the above markers, in AML cases: 1) CD14 FITC,
CD64PE, CD15 PE-Cy7, CD56 APC, CD45 APC-H7; 2) CD38 FITC, CD11c PE,
CD123 PerCP Cy5.5, CD2 PE-Cy7, CD11b APC, CD45 APC-H7 were used for
further characterizing Monocytic lineage AMLs and/or 3) CD235a FITC,
cytoplasmic CD61 PE, cytoplasmic CD41a PerCP Cy5.5, CD71 APC and CD45
APC-H7 were used in suspected erythroleukemia (AML M6) and
megakaryoblastic leukemia cases (AML M7). For B-ALLs, no further
markers were used. For T-ALL cases: 1) CD8 FITC, CD4 PE, CD99 PerCP
Cy5.5, CD2 PE-Cy7, CD11b APC, CD45 APC-H7; 2) CD57 FITC, CD16 PE, CD38
PerCP Cy5.5, sCD3 PE-Cy7, CD56 APC, CD45 APC-H7 were used for further
characterization. All antibodies mentioned above were obtained from
Becton Dickinson, San Jose, CA (USA).
Cytogenetics.
Bone marrow samples were processed using standard cytogenetic
protocols. For each sample, 15 – 20 GTG banded (G banding with trypsin
using Leishman stain) metaphases were obtained from at least two
un-stimulated overnight cultures of bone marrow (with and without
colcemid). An automated karyotyping system (MetaSystems GmbH,
Altlussheim, Germany) was used for analysis. Karyotypes were reported
in accordance with the International System for Human Cytogenetic
Nomenclature (ISCN) 2008.
FISH analysis (where applicable) was
performed for confirmation of translocations using dual color dual
fusion probes for BCR-ABL1 (Zytovision, Bremerhaven, Germany), t(8;21)
(Metasystems, Germany) and t(15;17) (Abbot Vysis, Illinois, U.S.A.),
locus-specific dual color break apart probes for MLL and inv(16) (Abbot
Vysis Illinois, U.S.A.) and locus-specific dual color extra signal
ETV6/RUNX1 probe (Abbot Vysis Illinois, U.S.A.) according to
manufacturer’s instructions following standard techniques.[5]
Molecular testing.
DNA was extracted from BM/ PB samples using the Qiagen DNA Mini kit
(Qiagen, Hilden, Germany). The ITD (Internal tandem duplications) and
tyrosine kinase domain (TKD) containing regions of the FLT3 and exon 12
of the NPM genes were amplified using fluorescent labeled primers. The
size of the ITD/ NPM PCR product was determined by ABI 3500 DNA
analyzer (Applied Biosystems, Foster City, California, USA). The TKD
PCR product was digested with EcoRV, and the presence of the mutation
was determined on agarose gel electrophoresis. RNA was extracted from
WBCs using Trizol reagent, complementary DNA was then synthesized using
reverse transcriptase, followed by nested RT-PCR (PML-RARA) using Roche
First Start Taq polymerase and the products were separated by agarose
gel electrophoresis.[6]
Results
Six
hundred thirty one newly diagnosed cases of AL were retrospectively
evaluated, of which 52.9% (n=334) were ALL, 43.9% (n=277) AML, 2.2%
(n=14) MPAL, 0.5% (n=3) acute undifferentiated leukemia (AUL) and 0.5%
(n=3) chronic myelogenous in blast crisis (CML-BC). Overall males
out-numbered females (M: F ratio, 1.5:1). Adults (≥ 15 years)
constituted 58.7 % (n=375) patients while 41.3% (n=256) belonged to
pediatric (<15 years) age group. AML was the commonest (59.7%,
n=224/375) AL in adults, while ALL was the commonest (59.0%, n=197/334)
in pediatric age group (Table 1).
Based on WHO 2008 defined categories, the immunophenotypic expression
pattern in all AL cases except T-ALL is depicted in Table 2 and that of T-ALL cases in Table 3.
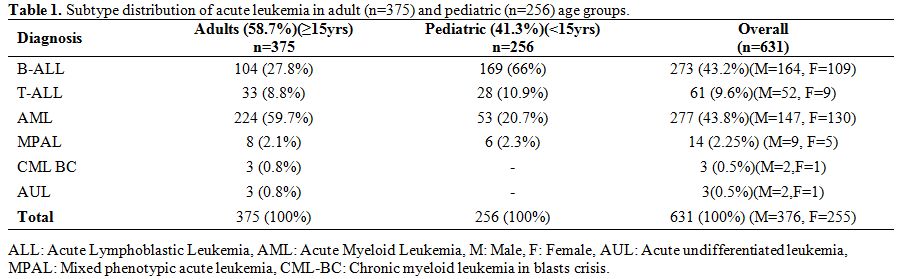 |
Table 1.
Subtype distribution of acute leukemia in adult (n=375) and pediatric (n=256) age groups. |
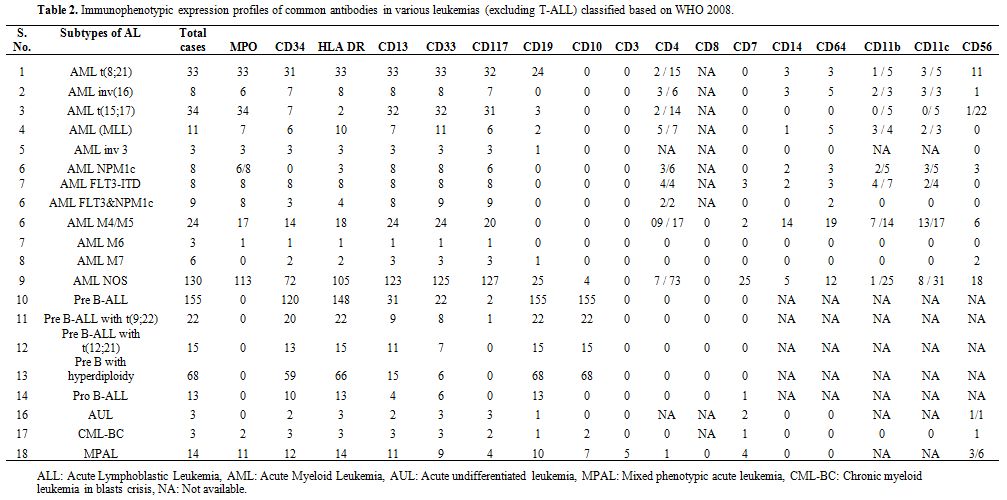 |
Table 2. Immunophenotypic
expression profiles of common antibodies in various leukemias
(excluding T-ALL) classified based on WHO 2008. |
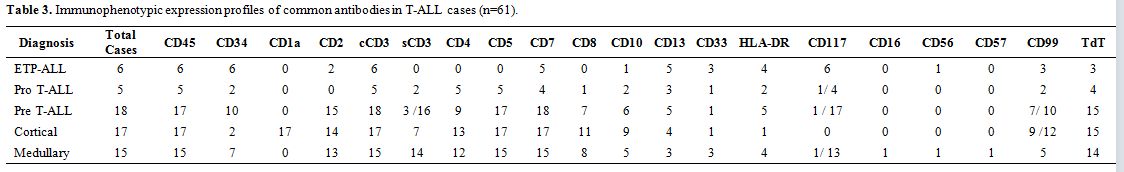 |
Table 3. Immunophenotypic expression profiles of common antibodies in T-ALL cases (n=61). |
Acute
lymphoblastic leukemia. Of the total 334 ALL cases, 81.7% (n=273) were
B-cell ALL and 18.3% (n=61) were T-cell ALL. B-cell ALLs included
common-B ALL (95.2%, n=260) and CD10 negative Pro-B ALL (4.8%, n=13)
cases (Table 2). Immaturity
markers like HLA-DR, TdT, and CD34 were expressed in 97.4%, 97% and
81.3% of cases respectively. CD13 was the commonest aberrantly
expressed marker in B-cell ALL, seen in 25.6% (n=70/273) cases followed
by CD33 in 17.9% (n=49/273) cases. CD13/CD33 co-expression was seen in
11.3% (n=31/273) cases (Table 4).
Cytogenetic data was available in 249 cases of which 15.7% (n=39) had a
normal karyotype. Remaining cases showed numerical/ structural
abnormalities or chromosomal translocations. Overall, 24.9% (n=68)
cases of B-ALL revealed hyperdiploidy of which 97% (n=66/68) belonged
to the pediatric age group. CD13 and CD33 were frequent aberrancies
observed in 22.1 % (n=15/68) and 8.8% (n=6/68) hyperdiploid cases
respectively, while 4.4% (n=3/68) co-expressed CD13/CD33. Philadelphia
positivity was found in 8.8% (n=22/249) cases, of which the majority
(86.4%, n=19/22) were adults. CD13 and CD33 aberrancies were noted in
40.9% (n=9/22) and 36.9% (n=8/22) cases respectively while CD13/ CD33
co-expression was seen in 31.8% (n=7/22) cases. Total 5.5% (n=15/270)
cases showed t(12;21) by FISH and CD13 was the most frequently (73.3%,
n=11/15) aberrantly expressed marker in this subgroup of patients also.
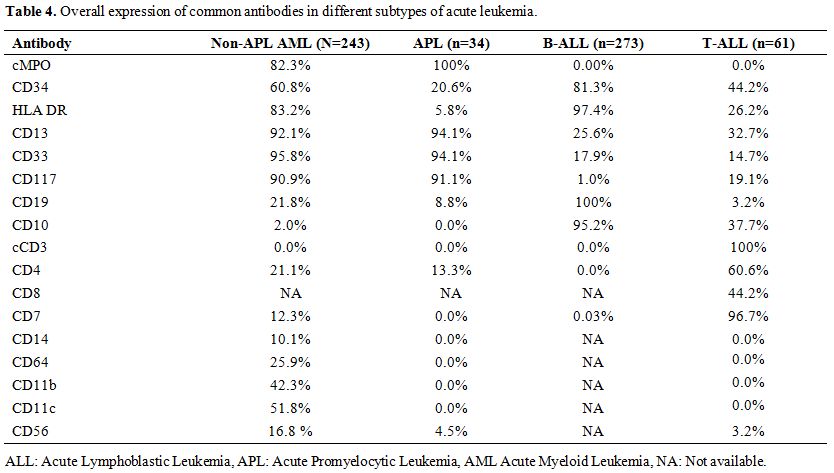 |
Table 4. Overall expression of common antibodies in different subtypes of acute leukemia. |
T-ALL
subgroup (n=61) included 9.8% (n=6) early thymic precursor T-ALL
(ETP-ALL), 8.2% (n=5) pro T-ALL, 29.5% (n=18) pre T-ALL, 27.9% (n=17)
cortical T-ALL and 24.6% (n=15) medullary T-ALL (Table 3).
All the cases of T-cell ALL consistently expressed cCD3. CD10 was
expressed in 37.7% (n=23/61) cases. CD13 was aberrantly expressed in
32.7% (n=20/61) cases and CD117 in 19.1% (n=9/47) cases (which were
positive for CD13 also in 88.8%, n=8/9 cases). CD4/CD8 co-expression
was seen in 39.3% (n=24/61) (Table 4).
Cytogenetic analysis was carried out in 51 cases (with n=7 culture
failures), of which 47.7% (n=21/44) had a normal karyotype. One case
had a complex karyotype while remaining (n=22) cases revealed various
structural and numerical abnormalities and balanced and unbalanced
translocations with unknown prognostic significance.
Acute myeloid leukemia.
A total of 277 cases were diagnosed as AML by flow cytometry and
further sub-classified based on the cytogenetic profiles. AML with
recurrent cytogenetic abnormalities constituted 32.1% (n=89/277) cases
including 11.9% (n=33/277) AML with t(8;21); 12.3% (n=34/277) AML with
t(15;17) i.e. APLs; 2.9% (n=8/277) AML with inversion 16/ t(16;16);
3.9% (n=11/277) MLL gene rearranged AMLs; and 1.1% (n=3/277) cases of AML with 3q abnormalities. Normal Karyotype AMLs with mutated NPM1/FLT3
constituted 9.0% (n=25/277) cases. Rest of the cases were categorized
into AML NOS (58.8%, n=163/277) which included 8.6% (n=24/277) cases of
AML with monocytic differentiation, 1.1% (n=3/277) acute erythroid
leukemias and 2.2% (n=6/277) acute megakaryoblastic leukemias (Table 2).
Flow
cytometric evaluation of all non-APL cases as a group (243/277) showed
consistent expression of CD33, CD13, CD117 and MPO in 95.8%, 92.1%,
90.9% and 82.3% of cases respectively. CD34 positivity was noted in
60.8 % of cases while HLA-DR was expressed in 83.2 % of non-APL AMLs.
Other markers like CD11b, CD11c, and CD64 were expressed in 42.3%,
51.8% and 25.9% of cases respectively. CD19 was the commonest
aberrantly expressed marker seen in 21.8% (n=53/243) of cases followed
by CD56 (16.8%, n=41/243) and CD7 (12.3%, n=30/243). Amongst 33 cases
with t(8;21), aberrant CD19 expression on flow cytometry was seen in
72.7% (n=24/33) of cases.
In APLs (n=34), MPO expression was seen
in all cases (100%) and CD13 and/or CD33 in 94.1% (n=32/34) of cases.
CD34 and HLA-DR were expressed in 20.6% (n=7/34) and 5.8% (n=2/34) of
cases respectively. CD11b and CD11c were analyzed in 5 cases of APL and
found to be consistently negative in all 5 cases (100%). We also found
aberrant CD4 expression in 13.3% (n=2/15) cases of APL and unusual
aberrant CD19 expression in 8.8% (n=3/34) of cases. In our cohort of
total n=34 APLs, RT-PCR PML/RARA analysis was available in n=24 cases,
of which 54.2% (n=13/24) had BCR1 transcript (all cases were CD34neg/CD56neg), 4.2% (n=1/24) had BCR2 (CD34neg/CD56neg)
and 41.2% (n=10) had BCR3 transcript (only n=1/10 case revealed CD34
positivity, while only one case had CD56 positivity). Conventional
karyotyping was available in n=33/34 cases, of which n=29/33 cases
showed t(15;17) on conventional karyotyping and were also FISH PML/RARA
positive. In the remaining five APL cases, FISH PML/RARA was negative
and karyotype (which was available in 4/5 cases) did not reveal any
structural abnormality. All these 5 cases were found to be RT-PCR
PML/RARA (BCR1 transcript) positive. We did not have any patients with
variant APML translocations in this cohort.
Amongst the cases of AML with normal karyotype (20.6%, n=57/277), FLT3/NPM
mutation analysis was carried out in 44 cases and found positive in
56.8% (n=25/44) cases. Eight of these cases (32%) were only NPM1c
mutated (all n=8/8 were CD34 negative, and n=6/8 were HLA-DR positive),
32% (n=8) were only FLT3 ITD mutated (all 8 were CD34 and HLA-DR
positive) and 36% (n=9) were both FLT3 ITD/ NPM1c mutated (showing CD34
expression in n=3/9 cases and HLA-DR in n=3/9 cases). Amongst the cases
tested, FLT3/ NPM1 mutations were not found in any of AML case with
recurrent cytogenetic abnormalities.
Acute Leukemia of ambiguous lineage.
Of the total 631 cases studied, 2.7% (n=17/630) cases were classified
as Acute Leukemia of Ambiguous lineage. Of these 17 cases, 82.4% (n=14)
cases were MPALs. Majority of these were males (57.1%, n=8) and adults
(78.5%, n=11). These cases were further classified as: B /myeloid
(64.3%, n=9/14), T / myeloid (28.6%, n=4/14) and B / T lymphoid (7.1%,
n=1/14). Cytogenetic data was available in 12 of 14 cases, of which 25%
(n=3) had normal karyotype and 33.3% (n=4) had t(9;22)(q34; q11).We
also came across a single case of B/T MPAL which had MLL (mixed lineage
leukemia) gene rearrangement, t(v;11q23). Certain cases (17.6%,
n=3/17), where none of the lineage-defining markers was expressed, were
classified into acute undifferentiated leukemia (AUL). All these cases
with treatment profiles have been already published earlier.[7]
CML in blasts crisis. CML in blast crisis constituted 3 cases. Two of these had myeloid lineage blast crisis and one lymphoid lineage (B-lymphoid).
Discussion
Flow
cytometric immunophenotyping is a highly sensitive and specific
technique and has become an integral part of diagnostic workup and
sub-classification of AL.[2] The immunophenotypic
profile also provides strong prognostic and predictive information. To
the best of our knowledge, published literature from the Indian
sub-continent on detailed AL immunophenotyping (including T-ALL) and
their corresponding surrogate marker profiles for genetic lesions is
sparse.
Total 631 consecutive cases of AL diagnosed at our center
were retrospectively evaluated. The overall patient demographic profile
and the frequency distribution of various AL subtypes were concordant
with published Indian and western literature.[8-17]
B-ALL immunophenotypic features and genetic findings.
B-ALL is defined by the presence of either strong CD19 along with one
other strongly expressed B- cell markers (i.e., CD79a, cCD22, CD10); or
weak CD19 and two other strongly expressed B-cell markers.[2] In our cohort, the overall B-ALL immunophenotypic profile was similar to that previously reported in the literature.[18-23]
Aberrant myeloid antigen (MyAg) expression has been reported to range from 4.3% to 64% in B-ALL.[24]
There is no reported difference between MyAg+ and MyAg− groups in
complete remission (CR) achievement or Overall Survival (OS).[20,24,25]
In our series, CD13 was the commonest aberrantly expressed marker in
B-cell ALL, seen in 25.6% (n=70/273) cases followed by CD33 in 17.9%
(n=49/273) cases while CD13/CD33 co-expression was seen in 11.3%
(n=31/273) cases. The cryptic (12;21)(p13;q22) translocation (TEL/AML1
fusion) found in > 20% pediatric ALL and < 1–3 % of adult ALLs
carries a favorable prognosis, particularly in pediatric population.[3,22]
Borkhardt et al. in their study on 334 pediatric ALLs showed that the
overall incidence of the t(12;21) was 18.9% and 24.6% cases
co-expressed at least two myeloid antigens (CD13, CD33, or CD65) in
more than 20% of the gated blast cells.[26] The most
predictive immune profile described in the literature for this
favorable risk subtype applicable for both pediatric and adult
population is CD10Pos, CD20Neg, CD34Neg, cIgMNeg, frequently CD33 and CD13Pos and CD11bNeg.[3]
In our study, t(12;21) was seen in 5.5% (n=15/270) cases (all
pediatric). CD13 was the most frequent (73.3%, n=11/15) aberrantly
expressed marker in this subgroup of patients followed by CD33 (46.6%,
n=7/15) and CD13/CD33 co-expression (33.3%, n=5/15). Our numbers for
t(12;21) positive cases were small in this cohort, and we did not have
cIgM and CD11b in our B-ALL panel so these markers could not be
evaluated.
BCR/ABL fusion gene is a dominant negative prognostic factor in ALL.[3,20] The best surrogate marker profile described in literature for BCR/ABLPos lymphoblasts includes CD25Pos / CD34high / CD10high / CD66cPos / CD38weak
blasts with dual expression of myeloid antigens CD33 and CD13.3 CD25
(alpha-chain of the interleukin-2 receptor) expression has been
described as an independent prognostic factor having negative impact on
OS, Event free survival (EFS) and low rates of CR in.[3,27-29]
In our cohort, Philadelphia positivity was found in 8.8% (n=22/249)
cases, of which majority i.e. 86.4% (n=19) were adults. CD13 and CD33
aberrancies were noted in 40.9% (n=9/22) and 36.9% (n=8/22) cases
respectively while CD13/ CD33 co-expression was seen in 31.8% (n=7/22)
cases. CD34 was positive in 90.9% (n=20/22) cases while 100% (n=22/22)
cases were CD10pos. We however, did not have CD25 and CD66c, in our diagnostic B-ALL panel.
CD10
(also known as the common acute lymphoblastic leukemia antigen, CALLA)
negativity in B-ALL (i.e., Pro B-ALL cases) is a strong negative
prognostic factor, frequently associated with MLL rearrangements and
are known to have a significantly low OS, EFS and low rates of CR.[3,22,24,27,28] In our series, 4.8% (n=13/273) of all B-ALLs were CD10neg.
CD20
expression, other than serving as a potential target for immunotherapy,
has known prognostic implications and is associated with shorter
remission duration and OS particularly in adults.[3] In our cohort, CD20 was found to be positive in 53.5% (n=146/273) cases similar to that reported in Indian literature[8-10]
but higher than that reported in western literature (20-40%). These
differences could be due to the choice of fluorochromes used in
different studies.[30]
T-ALL immunophenotypic features and genetic findings. T-lineage ALL is diagnosed based on the presence of cytoplasmic CD3 in leukemic blasts.[2]
Traditionally recognized T-ALL subsets based on different stages of
intra-thymic differentiation include pro-T ALL EGIL T-I (cCD3pos, CD7pos); pre-T ALL EGIL T- II (cCD3pos, CD7pos and CD5/CD2pos); cortical T ALL EGIL T-III (cCD3pos, CD1apos, sCD3pos/neg); and mature-T ALL EGIL T-IV (cCD3pos, sCD3pos, CD1aneg).[31-33]
In our cohort of T-ALLs (n=61/631), there were 8.2% (n=5) Pro T-cell
ALL, 29.5% (n=18) Pre T-ALL, 27.9% (n=17) Cortical T-ALL and 24.6%
(n=15) Medullary T- ALLs.
ETP-ALL (Early T-cell precursor ALL) is
a recently identified subtype of T-ALL characterized by distinctive
immunophenotype, gene expression profile (lower frequencies of
prototypical T-ALL lesions such as CDKN2A/B deletions, activating
mutations in NOTCH-signalling pathway, higher prevalence of mutations
typically associated with the pathogenesis of AMLs like NRAS/ KRAS/
FLT3), poor response to chemotherapy and very high risk of relapse.
ETP-ALLs show a uniquely high prevalence of 5q, 13q, and 11q
chromosomal deletions and in contrast, lack deletions involving the
short arm of chromosome 9 which delete the CDKN2A/B
tumor suppressor gene in over 70% of all T-ALLs. ETP-ALL
immunophenotype resembles the earliest thymic precursors with both T
and myeloid lineage potential.[31-34] Corroborating
with this truly bi-phenotypic potential, around 5-10% blasts in these
cases express cMPO and occasionally can show Auer rods, the majority of
these cases being FLT3 mutated.[3] In our series, 9.8%
(n=6/61) cases were classified into ETP-ALL based on the typical
immunophenotype characterized by expression of cCD3 (100% cases), CD7
(83.3% cases), TdT (50% cases), CD99 (50% cases) along with one or more
stem cell/ myeloid markers like CD34 (100% cases), CD117 (100% cases),
HLA-DR (66.6% cases), CD13 (83.3% cases), CD33 (50% cases) associated
with lack of CD1a (100% cases), CD8 (100% cases) and weak CD5 (100%
cases). ETP-ALLs also show significantly lower expression of CD2, sCD3,
CD4 and CD10,3 as seen in 33.3%, 0%, 0% and 16.6% of our cases
respectively. One of our ETP-ALL patients also had cMPO positivity in
5% blasts and showed occasional blasts with Auer rods, possibly hinting
towards FLT3 mutated ETP-ALL, which was not tested in any of the
patients with T-ALL. This immuno-morphological correlation in FLT3
mutated cases has implications concerning innovative treatment
strategies like tyrosine kinase inhibitors (TKI) for this otherwise
high-risk subgroup of adult ETP-ALLs.[3]
CD1a
antigen is expressed on 60-90% thymocytes, some T-cell leukemias/
lymphomas, Langerhans cells of the skin and some dendritic cells. Based
on CD1a along with CD13, from a prognostic point of view, particularly
in the adult population, two major subclasses of T-ALL which exhibit
minimal immunophenotypic overlap are relevant: CD1aPos Cortical/ Thymic T-ALL subtype (lacking CD13) and CD1aneg (CD13Pos CD34Pos
positive) T-ALL. Positivity of blasts for CD1a (lacking CD13
expression) has been associated with significantly higher OS, CR rate
and low risk of relapses compared with the CD1aNeg
(CD13 expressing) T-ALL. Overall, negativity for CD1a, positivity for
CD34, and the presence of myeloid antigens combined confer inferior
prognosis in T-ALL of all ages.[31,35]
Of
our total 61 T-ALL cases, CD13 was the most common aberrantly expressed
myeloid marker seen in 32.7% cases followed by CD33 (14.8% cases). In
the study by Marks DI et al, aberrant myeloid antigen expression i.e.
CD13 was seen in 51% and CD33 in 30% of T-ALL patients.[31]
Expression of CD10 and CD34 was observed in 34.4% and 32.7% of our
cases respectively. Published literature mentions expression of CD10 in
approximately 15-22% cases and CD34 in 10 to 42% T-ALLs.[18,19,21,32,33] In our series, CD4/CD8 co-expression was seen in 39.3% (n=24/61) cases, CD4/CD8 double negativity in 32.8% (n=20/61), CD4Pos/CD8Neg in 21.3% (n=13/61) and CD8Pos/CD4Neg in 6.6% cases (n=4) similar to that previously reported.[18,32,33]
Cytogenetic
analysis was carried out in 51 cases (n=7 cases with culture failure).
Of the remaining 44 cases, 47.7% (n=21/44) cases had a normal
karyotype. One case had a complex karyotype while remaining (n=22/44)
cases revealed various abnormalities including both euploidy and
aneuploidy. From a prognostic point of view, only patients with a
complex karyotype have been reported to have a significantly lower OS
compared with patients with normal karyotypes, irrespective of age and
WBC counts.[3,35]
AML subtypes with immunophenotypic features.
AML is diagnosed based on the presence of either myeloperoxidase (by
flow cytometry, immunohistochemistry or cytochemistry) or at least two
markers of monocytic differentiation (NSE, CD11c, CD14, CD64 or
lysozyme) in leukemic blasts.[2] Of our total 277 AML
cases (with n=243 Non APLs and n=34 APLs) expression of CD13, CD33 and
CD117 was seen in 93.9%, 94.8% and 90.9% respectively, similar to that
in published literature.[8,36-38]
In
our study, CD7 was aberrantly expressed in 10.8% of all AML cases
(n=30/277). CD7 is first to appear during the maturation of T
lymphocytes and is reported to be aberrantly expressed in an 11-28% of
AML cases.[9,13,36,39]
CD34, the stem cell marker, is known to be an independent negative
prognostic indicator for overall survival and remission achievement in
AMLs.[3,32] CD34 positivity in non-APL cases varies between 55.8 and 69.1%.[8,14,15] In our cohort, 60.8% of non-APL cases were CD34pos.
APL
was the first leukemia whose characteristic features were described
immunophenotypically. As per the ECOG guidelines, the most accepted
surrogate marker profile for PML/RARA includes lack of HLA-DR and CD133
(two antigens expressed at differentiation levels more immature than
that of promyelocytes during normal myelopoiesis); the absence of
adhesion molecules, such as CD11a (αL subunit of the leukocyte integrin
LFA-1), CD18 (β2 subunit of LFA-1), and CD11b (αMsubunit of Mac-1
integrin); the expression of carbohydrate molecule, CD15, only in the
sialylated form (CD15s); dim expression of CD45 and CD38 and strong
CD117 expression. These features of PML/RARA APL cells are valid for
all currently known variant APL translocations that involve
rearrangement of the RARA gene like the ATRA un-responsive PLZF/RARA
derived from t(11;17)(q23;q21), the ATRA responsive NPM/RARA
(nucleophosmin) APL, derived from t(5;17)(q35;q21) and also the FISH
negative, RT-PCR positive cases.[3,40]
In our cohort of APL cases, HLA-DR, CD34 and CD117 expression was seen
in 5.8%, 20.6%, and 91.1% cases respectively. Double negativity of CD34
and HLA-DR was seen in 77.1% of APL cases. Other markers like CD11c and
CD11b, tested in 5 out of our 34 cases, showed negativity in all 5
cases (100% negativity). Our findings are in concordance with published
literature.[8,13,16,17]
Based
on the differential breakpoints in the PML gene, the L- (Long, bcr1),
S- (Short, bcr3), or the V- (Variable, bcr2) transcript isoform of
PML/RARA are formed. The classical surrogate marker profile described
applies to all these three molecular isoforms. However, only the
leukemic promyelocytes that contain S-form transcripts variably express
CD34, CD2, and CD56 and are associated with a higher incidence of
extra-medullary relapse, greater thrombotic complications and poorer
prognosis.[3,40] These cases are
also associated with FLT3 mutations. CD2 and CD56 at diagnosis are
never seen in BCR1 and BCR2 PML/ RARA transcripts, however, can rarely
have CD34 expression.[3] In our cohort of patients, of
the total n=34 APLs, RT-PCR PML/RARA analysis was available in n=24
cases, of which 54.2% (n=13) had BCR1 transcript (all cases were CD34neg/CD56neg), 4.2% (n=1) had BCR2 (CD34neg/CD56neg)
and 41.2% (n=10) had BCR3 transcript (n=1 case revealed CD34 positivity
another case had CD56 positivity). We did not have CD2 in our
diagnostic panel so could not be evaluated.
The characteristic
feature of the t(8;21) AML immunophenotype is striking myeloid
immaturity, with weak expression of CD33 and weak MPO consistent with
the suggestion that the AML1 / ETO fusion event occurs at an early
stem/ progenitor cell stage AML. The unique B-lymphoid aberrant
antigenic signature of t(8;21) AML, with an expression of PAX5, CD19,
and CD79a is however negated by the co-expression with CD7, and the
CD19/CD7 double positivity has been associated with a predominantly
normal karyotype and FLT3-ITD and NPM1 mutations.[3]
Aberrant expression of CD19 was noted in 20.2% (n=56/277) AML cases in
our cohort. It has been reported in 2-22% AMLs in literature, being
relatively more frequent in AML with translocation t(8;21).[41]
In our study, of the 33 cases of AML with t(8;21), CD19 aberrancy was
seen in 72.7% (n= 24/33) cases while none of these cases (100%, n=24)
had CD7 aberrant expression along with CD19. In the study by Gujral et
al., CD19 aberrancy predicted t(8;21)(q22;q22) in 45% of cases.[8] CD56 (neural cell adhesion molecule) is associated with increased incidence of granulocytic sarcomas in t(8;21) AMLs.[3,37,39,41]
In our cohort, 33.3% (n=11/33) cases revealed aberrant CD56 expression
of which eight presented with the extra-medullary disease.
AML
patients with normal cytogenetics are characterized by CD34 and HLA-DR
negativity. These cases may carry poor prognostic genetic lesions (FLT3
mutations) or aberrations associated with better outcome (isolated NPM1
or CEBPA mutations). The distinctive “cuplike” nuclei (nuclear
invaginations) are typically associated with AMLs having Normal
Karyotype and mutated NPM1 and/or FLT3. NPM1/ FLT3 mutations can be
seen irrespective of FAB AML subtypes. NPM1 mutations involve all but
lymphoid hematopoietic cell lineages. However, FLT3 mutations can also
be seen in T-ALL (more commonly in pediatric patients) and very rarely
also seen in B-ALL.[3,42] In AML,
CD34 expression is related to genes predictive of poor outcome (FLT3),
while a lack of CD34 expression predicts favorable prognosis. In our
cohort of AMLs with normal karyotype (20.6%, n=57/277), FLT3/NPM
mutation analysis was carried out in 44 cases and found positive in
56.8% (n=25/44) cases. Eight of these cases (32%) were only NPM1c
mutated (all eight were CD34 negative, and only 6/8 were HLA-DR
positive), 32% (n=8) were only FLT3 ITD mutated (all 8 were CD34 and
HLA-DR positive) and 36% (n=9) were both FLT3 ITD/ NPM1c mutated
(showing CD34 expression in 3/9 cases and HLA-DR in 3/9 cases).
Mixed phenotypic leukemia.
MPALs are thought to arise from multipotent progenitor stem cells
capable of differentiating into both myeloid and lymphoid lineages.[7,43-45]
The frequency of MPAL in our study was found to be 2.2%
(n=14/631) which is in concordance with published data, with
documented frequency ranging from 2.2 to 2.6%.[43,44]
CD34 expression was seen in 85.7% (n=12) cases. MPAL were classified
into B/myeloid in 64.2% (n=9/14), T/Myeloid in 28.5% (n=4/14) along
with a single case of B+T lymphoid MPAL based on WHO 2008 criteria. A
similar study by Matutes E, et al. consisting of a larger cohort of 100
MPAL, diagnosed using 2008 WHO criteria, revealed 59% of cases were
B/myeloid, 35% T/myeloid and B+T lymphoid included 4% cases.[43]
Myeloid markers frequently co-expressed with MPO in our study included
CD13/CD33 (69%) and CD117 (38%). Matutes et al. in their study found
that majority of lymphoid/ myeloid MPALs frequently expressed myeloid
markers like CD13 (74%), CD33 (66%), and CD117 (52%) along with MPO.[43] A description of MPAL cases in our cohort has been published recently.[7]
Conclusions
In
this study, we document the spectrum, correlate the immunophenotype
with genetic data of all leukemias, especially with respect to T-ALL
where the data from Indian sub-continent is scarce.
Acknowledgement
We would like to acknowledge the clinical hematology, pediatric oncology, hemato-pathology, and cytogenetics team.References
- Muniraj F. Classification of Acute Leukemias – Past, Present and Future. IJSS Case Reports & Reviews 2015;1(12):61-66.
- Swerdlow
S, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman
JW. WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues 4th Ed.(2008).
- Paietta E. Immunobiology of acute leukemia. In Neoplastic Diseases of the Blood 2013 (pp. 241-283). Springer New York. https://doi.org/10.1007/978-1-4614-3764-2_17
- Bain,
Barbara J., Imelda Bates, and Mike A. Laffan. Dacie and Lewis Practical
Haematology E-Book. Elsevier Health Sciences, 2016.
- Korf BR. Overview of clinical cytogenetics. Current Protocols in Human Genetics. 2001 Aug:8-1.
- Leonard
DG, Bagg A, Caliendo AM, Deerlin VM, Kaul KL, editors. Molecular
pathology in clinical practice. Springer Science+ Business Media, LLC;
2007 Nov 25. https://doi.org/10.1007/978-0-387-33227-7
- Pawar
RN, Banerjee S, Bramha S, Krishnan S, Bhattacharya A, Saha V,
Chakrapani A, Bhave S, Chandy M, Nair R, Parihar M. Mixed-phenotypic
acute leukemia series from tertiary care center. Indian Journal of
Pathology and Microbiology. 2017 Jan 1;60(1):43. PMid:28195090
- Gujral
S, Badrinath Y, Kumar A, Subramanian PG, Raje G, Jain H, Pais A, Kadam
PS, Banavali SD, Arora B, Kumar P. Immunophenotypic profile of acute
leukemia: critical analysis and insights gained at a tertiary care
center in India. Cytometry Part B: Clinical Cytometry. 2009 May
1;76(3):199-205. https://doi.org/10.1002/cyto.b.20451 PMid:18803279
- Ghosh
S, Shinde SC, Kumaran GS, Sapre RS, Dhond SR, Badrinath Y, Ansari R,
Kumar A, Mahadik S, Chougule AB, Nair CN. Haematologic and
immunophenotypic profile of acute myeloid leukemia: an experience of
Tata Memorial Hospital. Indian Journal of Cancer. 2003 Apr 1;40(2):71.
PMid:14716122
- Advani S, Pai S, Venzon D,
Adde M, Kurkure PK, Nair CN, Sirohi B, Banavali SD, Hawaldar R,
Kolhatkar BB, Vats T. Acute lymphoblastic leukemia in India: an
analysis of prognostic factors using a single treatment regimen. Annals
of oncology. 1999 Feb;10(2):167-76. https://doi.org/10.1023/A:1008366814109 PMid:10093685
- El
Sharkawy N, Abdel Hamid T. Internal tandem duplication of FLT3 gene in
Egyptian adult acute myeloid and acute lymphoblastic leukemia. J Am
Sci. 2010;6(9):14-22.
- Salah SM. Detection of CXCL12 gene polymorphism and CXCR4 receptor expressionin Egyptian acute myeloid leukemia patients.
- Salem
DA, El-Aziz SM. Flowcytometric immunophenotypic profile of acute
leukemia: mansoura experience. Indian journal of hematology and blood
transfusion. 2012 Jun 1;28(2):89-96. https://doi.org/10.1007/s12288-011-0110-2 PMid:23730015 PMCid:PMC3332273
- Nakase
K, Bradstock K, Sartor M, Gottlieb D, Byth K, Kita K, Shiku H, Kamada
N. Geographic heterogeneity of cellular characteristics of acute
myeloid leukemia: a comparative study of Australian and Japanese adult
cases. Leukemia. 2000 Jan 1;14(1):163. https://doi.org/10.1038/sj.leu.2401638 PMid:10637492
- Chang
H, Salma F, Yi QL, Patterson B, Brien B, Minden MD. Prognostic
relevance of immunophenotyping in 379 patients with acute myeloid
leukemia. Leukemia Research. 2004 Jan 31;28(1):43-8. https://doi.org/10.1016/S0145-2126(03)00180-2
- Wang
XB, Zheng JE, Gu JX, Yao JX, Yang J, Liu J, Li XQ, He YL, Yu JM, Wei J,
Liu ZP. Correlation of immunophenotype to cytogenetics and clinical
features of adult acute myeloid leukemia. Ai zheng= Aizheng= Chinese
Journal of Cancer. 2005 Jun;24(6):667-71.
- Dong
HY, Kung JX, Bhardwaj V, McGill J. Flow cytometry rapidly identifies
all acute promyelocytic leukemias with high specificity independent of
underlying cytogenetic abnormalities. American Journal of Clinical
Pathology. 2011 Jan 1;135(1):76-84. https://doi.org/10.1309/AJCPW9TSLQNCZAVT PMid:21173127
- Bachir
F, Bennani S, Lahjouji A, Cherkaoui S, Khattab M, Nassereddine I, Zafad
S, El Aouad R. Characterization of acute lymphoblastic leukemia
subtypes in Moroccan children. International Journal of Pediatrics.
2009 Jul 19;2009.
- Bhushan B, Chauhan PS,
Saluja S, Verma S, Mishra AK, Siddiqui S, Kapur S. Aberrant phenotypes
in childhood and adult acute leukemia and its association with adverse
prognostic factors and clinical outcome. Clinical and Experimental
Medicine. 2010 Mar 1;10(1):33-40. https://doi.org/10.1007/s10238-009-0067-8 PMid:19779962
- Vitale
A, Guarini A, Ariola C, Meloni G, Perbellini O, Pizzuti M, De Gregoris
C, Mettivier V, Pastorini A, Pizzolo G, Vignetti M. Absence of
prognostic impact of CD13 and/or CD33 antigen expression in adult acute
lymphoblastic leukemia. Results of the GIMEMA ALL 0496 trial.
Haematologica. 2007 Mar 1;92(3):342-8. https://doi.org/10.3324/haematol.10385 PMid:17339183
- Shen
HQ, Tang YM, Yang SL, Qian BQ, Song H, Shi SW, Xu WQ. Immunophenotyping
of 222 children with acute leukemia by multi-color flow cytometry.
Zhonghua er ke za zhi. Chinese Journal of Pediatrics. 2003
May;41(5):334-7. PMid:14751050
- Woo JS,
Alberti MO, Tirado CA. Childhood B-acute lymphoblastic leukemia: a
genetic update. Experimental hematology & oncology. 2014 Jun
13;3(1):16. https://doi.org/10.1186/2162-3619-3-16 PMid:24949228 PMCid:PMC4063430
- Velizarova
M, Popova D, Hadjiev E, Aleksandrova K, Dimova I, Zaharieva B, Toshkov
S, Staneva M, Hodjajik D, Penev M, Toncheva D. Molecular-cytogenetic
aberrations in B-cell adult acute lymphoblastic leukemia
(B-ALL)-frequency and correlation with immunophenotype. Turk J Hematol.
2006 Sep 5;23:151-7. PMid:27265483
- Supriyadi
E, Veerman AJ, Purwanto I, Cloos J. Myeloid antigen expression in
childhood acute lymphoblastic leukemia and its relevance for clinical
outcome in indonesian ALL-2006 protocol. Journal of Oncology. 2012 Nov
26;2012.
- Seegmiller AC, Kroft SH,
Karandikar NJ, McKenna RW. Characterization of immunophenotypic
aberrancies in 200 cases of B acute lymphoblastic leukemia. American
Journal of Clinical Pathology. 2009 Dec 1;132(6):940-9. https://doi.org/10.1309/AJCP8G5RMTWUEMUU PMid:19926587
- Borkhardt
A, Cazzaniga G, Viehmann S, Valsecchi MG, Ludwig WD, Burci L, Mangioni
S, Schrappe M, Riehm H, Lampert F, Basso G. Incidence and clinical
relevance of TEL/AML1 fusion genes in children with acute lymphoblastic
leukemia enrolled in the German and Italian multicenter therapy trials.
Blood. 1997 Jul 15;90(2):571-7. PMid:9226156
- Corrente
F, Bellesi S, Metafuni E, Puggioni PL, Marietti S, Ciminello AM, Za T,
Sorà F, Fianchi L, Sica S, De Stefano V. Role of flow‐cytometric
immunophenotyping in prediction of BCR/ABL1 gene rearrangement in adult
B‐cell acute lymphoblastic leukemia. Cytometry Part B: Clinical
Cytometry. 2018 May;94(3):468-76. https://doi.org/10.1002/cyto.b.21605 PMid:29220871
- Tabernero
M, Bortoluci AM, Alaejos I, Lopez-Berges MC, Rasillo A, Garcia-Sanz R,
Garcia M, Sayagues JM, Gonzalez M, Mateo G, San Miguel JF. Adult
precursor B-ALL with BCR/ABL gene rearrangements displays a unique
immunophenotype based on the pattern of CD10, CD34, CD13 and CD38
expression. Leukemia. 2001 Mar;15(3):406. https://doi.org/10.1038/sj.leu.2402060 PMid:11237064
- Yang
C, Yang LH, Zhang RJ, Ge XY, Wang MF, Ren FG, Zhang YF, Hou YF, Wang
YP. Expression of CD25 in acute B cell lymphoblastic leukemia and its
clinical significance. Zhongguo shi yan xue ye xue za zhi. 2014
Jun;22(3):634-9. PMid:24989267
- Kumar J,
Khan AA, Saraf A, Bhargava M. Expression of CD20 in B Cell Precursor
Acute Lymphoblastic Leukemia. Indian Journal of Hematology and Blood
Transfusion. 2014 Mar 1;30(1):16-8. https://doi.org/10.1007/s12288-012-0216-1 PMid:24554814 PMCid:PMC3921324
- Marks
DI, Paietta EM, Moorman AV, Richards SM, Buck G, DeWald G, Ferrando A,
Fielding AK, Goldstone AH, Ketterling RP, Litzow MR. T-cell acute
lymphoblastic leukemia in adults: clinical features, immunophenotype,
cytogenetics, and outcome from the large randomized prospective trial
(UKALL XII/ECOG 2993). Blood. 2009 Dec 10;114(25):5136-45. https://doi.org/10.1182/blood-2009-08-231217 PMid:19828704 PMCid:PMC2792210
- Basso
G, Lanza F, Orfao A, Moretti S, Castoldi G. Clinical and biological
significance of CD34 expression in acute leukemia. Journal of
Biological Regulators and Homeostatic Agents. 2001 Jan 1;15(1):68-78.
PMid:11388747
- Dakka N, Bellaoui H,
Bouzid N, Khattab M, Bakri Y, Benjouad A. CD10 AND CD34 expression in
childhood acute lymphoblastic leukemia in Morocco: clinical relevance
and outcome. Pediatric Hematology and Oncology. 2009 Jan
1;26(4):216-31. https://doi.org/10.1080/07357900902897557 PMid:19437324
- Jain
N, Lamb AV, O'Brien S, Ravandi F, Konopleva M, Jabbour E, Zuo Z,
Jorgensen J, Lin P, Pierce S, Thomas D. Early T-cell precursor acute
lymphoblastic leukemia/lymphoma (ETP-ALL/LBL) in adolescents and
adults: a high-risk subtype. Blood. 2016 Apr 14;127(15):1863-9. https://doi.org/10.1182/blood-2015-08-661702 PMid:26747249 PMCid:PMC4915808
- Chiaretti
S, Zini G, Bassan R. Diagnosis and subclassification of acute
lymphoblastic leukemia. Mediterranean Journal of Hematology and
Infectious Diseases. 2014;6(1). https://doi.org/10.4084/mjhid.2014.073 PMid:25408859 PMCid:PMC4235437
- Webber
BA, Cushing MM, Li S. Prognostic significance of flow cytometric
immunophenotyping in acute myeloid leukemia. International Journal of
Clinical and Experimental Pathology. 2008;1(2):124. PMid:18784805
PMCid:PMC2480555
- Legrand O, Perrot JY,
Baudard M, Cordier A, Lautier R, Simonin G, Zittoun R, Casadevall N,
Marie JP. The immunophenotype of 177 adults with acute myeloid
leukemia: proposal of a prognostic score. Blood. 2000 Aug
1;96(3):870-7. PMid:10910899
- Osman IM,
Humeida AA, Eltayeb O, Abdelrhman I, Elhadi TA. Flowcytometric
Immunophenotypic characterization of acute myeloid leukemia (AML) in
Sudan. International Journal of Hematological Disorders. 2015 Jan
23;2(1):10-7.
- Khalidi HS, Medeiros LJ,
Chang KL, Brynes RK, Slovak ML, Arber DA. The immunophenotype of adult
acute myeloid leukemia: high frequency of lymphoid antigen expression
and comparison of immunophenotype, French-American-British
classification, and karyotypic abnormalities. American Journal of
Clinical Pathology. 1998 Feb 1;109(2):211-20. https://doi.org/10.1093/ajcp/109.2.211 PMid:9583894
- Sainty
D, Liso V, Cantù-Rajnoldi A, Head D, Mozziconacci MJ, Arnoulet C,
Benattar L, Fenu S, Mancini M, Duchayne E, Mahon FX. A new morphologic
classification system for acute promyelocytic leukemia distinguishes
cases with underlyingPLZF/RARA gene rearrangements. Blood. 2000 Aug
15;96(4):1287-96. PMid:10942370
- Ortolani C. Flow cytometry of hematological malignancies. John Wiley & Sons; 2011 Aug 15.
- Papaemmanuil
E, Gerstung M, Bullinger L, Gaidzik VI, Paschka P, Roberts ND, Potter
NE, Heuser M, Thol F, Bolli N, Gundem G. Genomic classification and
prognosis in acute myeloid leukemia. New England Journal of Medicine.
2016 Jun 9;374(23):2209-21. https://doi.org/10.1056/NEJMoa1516192 PMid:27276561 PMCid:PMC4979995
- Matutes
E, Pickl WF, van't Veer M, Morilla R, Swansbury J, Strobl H,
Attarbaschi A, Hopfinger G, Ashley S, Bene MC, Porwit A.
Mixed-phenotype acute leukemia: clinical and laboratory features and
outcome in 100 patients defined according to the WHO 2008
classification. Blood. 2011 Mar 17;117(11):3163-71. https://doi.org/10.1182/blood-2010-10-314682 PMid:21228332
- Yan
L, Ping N, Zhu M, Sun A, Xue Y, Ruan C, Drexler HG, MacLeod RA, Wu D,
Chen S. Clinical, immunophenotypic, cytogenetic, and molecular genetic
features in 117 adult patients with mixed-phenotype acute leukemia
defined by WHO-2008 classification. Haematologica. 2012 Nov
1;97(11):1708-12. https://doi.org/10.3324/haematol.2012.064485 PMid:22581002 PMCid:PMC3487445
- Eckstein
OS, Wang L, Punia JN, Kornblau SM, Andreeff M, Wheeler DA, Goodell MA,
Rau RE. Mixed-phenotype acute leukemia (MPAL) exhibits frequent
mutations in DNMT3A and activated signaling genes. Experimental
hematology. 2016 Aug 1;44(8):740-4. https://doi.org/10.1016/j.exphem.2016.05.003 PMid:27208809 PMCid:PMC49565
[TOP]



