Ping Qiang1,2, Qing Pan3, Chao Fang4, Claudio Fozza5, Kaidi Song2, Yuanyuan Dai4, Wenjiao Chang4, Wei Chen6, Wan Yao7, Weibo Zhu2, Xin Liu2 and Xiaoling Ma1,4.
1
School of Medicine, Shandong University, Jinan, 250100, China.
2 Department of Hematology, The First Affiliated Hospital of University of Science and Technology of China, Hefei, 230001, China.
3 The First Hospital Affiliated to Anhui University of Traditional Chinese Medicine, Hefei, 230001, China.
4
Department of Laboratory Medicine, The First Affiliated Hospital of
University of Science and Technology of China, Hefei, 230001, China.
5 University of Sassari, Sassari, Italy.
6 School of Computer Science, University of Science and Technology of China, Hefei, 230001, China.
7 School of clinical medical, University of medical of Anhui, Hefei, 230001, China.
Corresponding
author: Prof. Xiaoling Ma, School of Medicine, Shandong University,
Jinan, China. Department of Laboratory Medicine, The First Affiliated
Hospital of University of Science and Technology of China, Hefei,
230001, China. Tel: +86-551-62283454, Fax: +86-551-62283454 E-mail:
maxiaoling@ustc.edu.cn
Published: March 1, 2020
Received: July 12, 2020
Accepted: February 3, 2020
Mediterr J Hematol Infect Dis 2020, 12(1): e2020012 DOI
10.4084/MJHID.2020.012
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Background:
Micro (mi) RNAs play an important role in the pathogenesis and
development of acute myeloid leukemia (AML), and their abnormal
expression may be sufficient to predict the prognosis and outcomes in
AML patients. We evaluated the clinical diagnostic value of
miRNA-181a-3p in predicting prognosis and outcomes in patients with AML.
Methods:
A total of 119 newly diagnosed adult patients with AML and 60 healthy
controls were recruited. Blood specimens were obtained from all AML
patients at diagnosis, and 10 blood specimens were obtained on day 28
after induction chemotherapy. The controls also provided blood samples.
Relative gene expression was quantified by PCR and determined using the
comparative Ct method. Publicly available clinical data and gene
expressions for 188 patients with AML were downloaded from TCGA data
portal.
Results: Compared
with healthy controls, the expression of miRNA-181a-3p was
significantly increased in patients with AML. MiR-181a-3p expression
could be used to discriminate AML patients from controls, with
up-regulated expression correlating with favorable prognosis. Moreover,
miRNA-181a-3p expression was significantly decreased in patients who
achieved a complete response after induction chemotherapy. The
multivariate Cox analysis highlighted the prognostic value of
miR-181a-3p for patients with AML. Finally, we found that miR-181a-3p
expression was negatively correlated with the expression of the NF-κB
essential modulator (NEMO/IKBKG).
Conclusions:
MiR-181a-3p may be clinically useful as a disease marker for AML, and
enhanced the prediction of patient outcomes to chemotherapy.
|
Introduction
Acute
myeloid leukemia (AML) is one of the most common adult leukemias. It is
a molecularly heterogeneous disease that is generally associated with
poor outcomes. AML patients are classified into distinct risk
categories for risk-adjusted chemotherapy, on the basis of cytogenetic
and molecular abnormalities.[1] Patients with complex
karyotype abnormalities or unfavorable molecular characteristics often
have an unfavorable prognosis. However, not all AML patients carry
cytogenetic alterations, so new genomic approaches to improve risk
stratification are needed.
Micro (mi)RNAs are small, noncoding RNAs that bind their target mRNAs and inhibit the expression of encoded proteins.[2]
MiRNAs have critical biological functions, including in hematopoietic
cell proliferation, differentiation, and apoptosis, and may also play
an essential role in the pathogenesis and development of AML.[3]
Several studies have identified that distinctive miRNA profiles are
associated with cytogenetic subtypes, mutations, and clinical outcomes
of AML.[3-5] For example, decreased miR-196b
expression is associated with the absence of FLT3-ITD and NPM1
mutations, and high miR-196b expression acts as a predictive factor of
poor prognosis.[4-5] Therefore, miRNA expression levels may be suitable to predict prognosis and outcomes in AML patients.
The
miR-181 family is thought to be involved in a number of biological
processes, including transcription, translation, and signaling
transduction.[6] In humans, the miR-181 family has four mature homologs (hsa-miR-181a, hsa-miR-181b, hsa-miR-181c and hsa-miR-181d).[7]
MiR-181a-3p belongs to miR-181a mature homologs, acts as a negative
post-transcriptional regulator of Nuclear Factor kappa-B (NF-κB)
signaling pathway by directly targeting NF-κB essential modulator
(NEMO/IKBKG) in Human Umbilical Vein Endothelial Cells (HUVECs).[8]
Our previous study showed that abnormal expression of miR-181a-3p was
associated with human monocytic leukaemia cell line THP-1 cell.[9]
However, few studies have focused on the clinical role of miR-181a-3p
in AML patients. Therefore, the present study examined the expression
of miR-181a-3p in AML patients prior to treatment to evaluate its
clinical diagnostic value and predictive role in the prognosis and
outcomes of AML patients.
Materials and Methods
RNA extraction, real-time PCR and miR-181a-3p Expression Analyses.
Total RNA was extracted from blood cells using TRIzol (Invitrogen).
Reverse transcription and quantitative real-time PCR (RT-qPCR) for gene
expression were performed using the SYBR Green PCR Kit (GenePharma,
shanghai, PR China). The primer sequences were as follows. MiR-181-a-3p
forward (5’-3’): AGAATTACACCATCGACCGTTG; MiRNA-181-a-3p reverse
(5’-3’): TATGCTTGTTCTCGTCTCTGTGTC. U6 forward (5’-3’):
ATTGGAACGATACAGAGAAGATT; U6 reverse (5’-3’): GGAACGCTTCACGAATTTG. NF-κB
forward (5’-3’): CTGAACCAGGGCATACCTGT; NF-κB reverse (5’-3’):
GAGAAGTCCATGTCCGCAAT. NEMO/IKBKG forward (5’-3’): TACTGGGCGAAGAGTCTCC;
NEMO/IKBKG reverse (5’-3’): AGAATCTGGTTGCTCTGCC. Analysis of relative
gene expression was using 2−△△CT method.
TCGA data set. Publicly available clinical, gene expression data for 188 patients with AML were downloaded from TCGA data portal.
Data analysis.
We used SPSS (version 24), GraphPad Prism (version 7) software and R
language to analyze the data. The unpaired Student’s t test
(two-tailed) was performed to compare differences in miRNA expression
between different groups. Chi-square tests were used to test the
association between the expression of miRNAs and clinicopathological
characteristics. Cox proportional hazards models were used to analyze
the prognostic utility of miRNA expression for disease-free survival
(DFS) and overall survival (OS) in AML patients.
Results
Patient clinical characteristics and treatment.
From September 2014 through December 2016, a total of 119 untreated,
newly diagnosed adult AML patients [age range 15-83 years; male, 52.1%]
in The First Affiliated Hospital of University of Science and
Technology of China were recruited , together with 60 heath controls
[age range 16-81 years; male, 57%] with no hematologic disease. The
patients represented the major French-American-British (FAB) subtypes:
1M0, 9 M1, 30 M2, 20 M3, 16 M4, and 43 M5. Of the 119 AML patients, 20
had PML-RARa rearrangements, 24 had AML1-ETO rearrangements, 30 had
other molecular genetic abnormalities, and 45 had normal karyotypes. 20
M3 patients received ARTA plus anthracycline-based induction
chemotherapy with or without ATO, and 99 AML patients received
traditional 7+3 induction chemotherapy. A total of 81 patients achieved
a complete response (CR), 30 did not, and 8 patients died within 30
days of receiving induction chemotherapy. The patients' clinical
characteristics were summarized in Table 1.
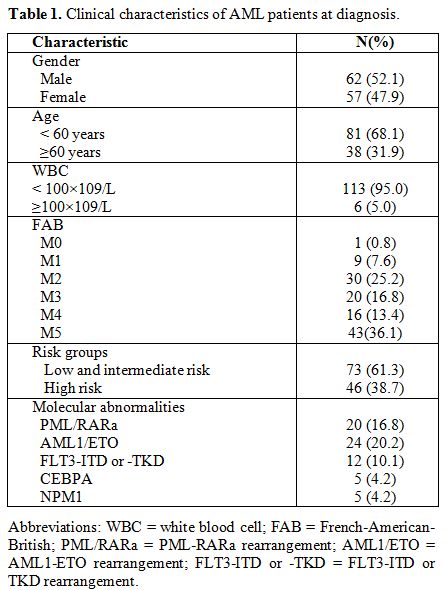 |
Table
1. Clinical characteristics of AML patients at diagnosis. |
The clinical value of miRNA-181a-3p in the diagnosis of AML.
To examine whether miRNA-181a-3p was abnormally expressed in patients
with AML, we detected miRNA expression in 60 healthy controls and 119
adult patients with newly diagnosed AML. Compared with healthy
controls, the expression of miRNA-181a-3p (P<0.001, Figure 1 A) was significantly increased in AML patients, and in the samples of M1, M2, M3, and M4 subtypes (Figure 1 B).
Furthermore, we compared the expression of microRNAs in 4 subtypes base
on molecular genetic abnormalities. Compared with healthy controls, the
expression of miRNA-181a-3p was significantly increased in all four
subtypes (Figure 1 C).
To
assess the clinical diagnostic value of miR-181a-3p in discriminating
AML patients from healthy controls, we performed receiver operating
characteristic (ROC) curve analyses. The Area Under Curve (AUC) of
miR-181a-3p was (0.654, 95% CI, 0.575 to 0.732, P<0.001, Figure 1D). Compared with health controls, miR-181a-3p showed significant difference in the samples of FAB M1, M2, M3 subtypes (Figure 1E) and PML/RARa, AML1/ETO subtypes (Figure 1F)
compared with controls, suggesting that it had value in discriminating
patients from controls. The AUC of miR-181a-3p in the samples of
different subtypes was showed in Table 2.
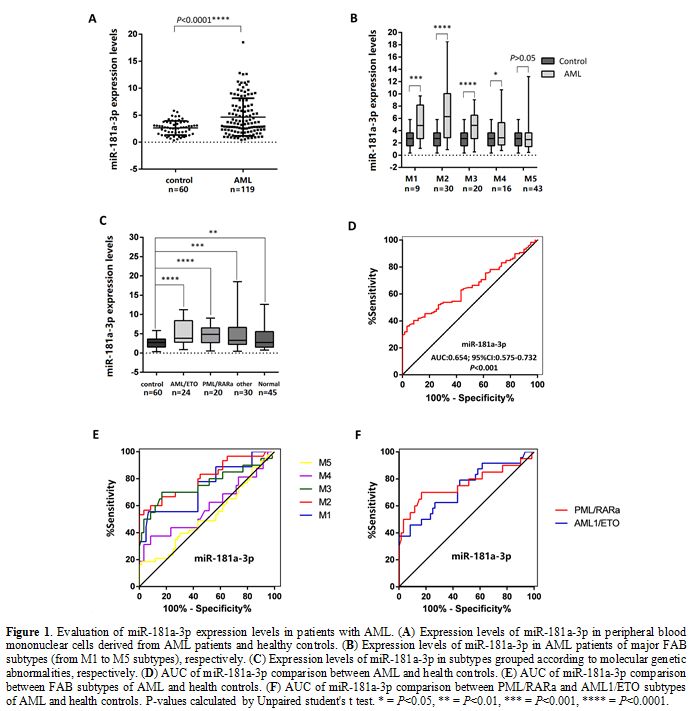 |
Figure 1 Evaluation of miR-181a-3p expression levels in patients with AML. (A) Expression levels of miR-181a-3p in peripheral blood mononuclear cells derived from AML patients and healthy controls. (B) Expression levels of miR-181a-3p in AML patients of major FAB subtypes (from M1 to M5 subtypes), respectively. (C) Expression levels of miR-181a-3p in subtypes grouped according to molecular genetic abnormalities, respectively. (D) AUC of miR-181a-3p comparison between AML and health controls. (E) AUC of miR-181a-3p comparison between FAB subtypes of AML and health controls. (F)
AUC of miR-181a-3p comparison between PML/RARa and AML1/ETO subtypes of
AML and health controls. P-values calculated by Unpaired student's t
test. * = P<0.05, ** = P<0.01, *** = P<0.001, **** =
P<0.0001. |
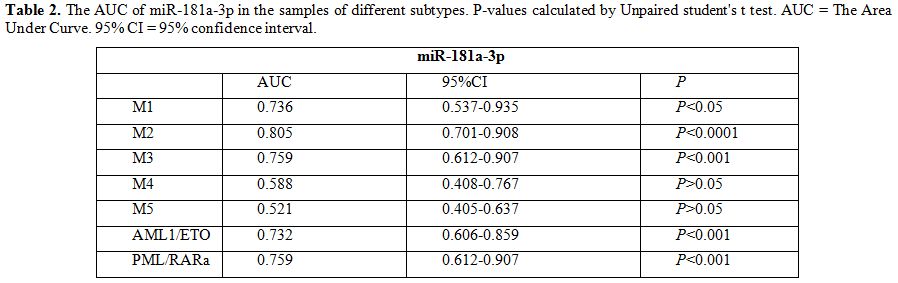 |
Table 2. The AUC of miR-181a-3p in the
samples of different subtypes. P-values calculated by Unpaired
student's t test. AUC = The Area Under Curve. 95% CI = 95% confidence
interval. |
The expression of miRNA-181a-3p was decreased when patients achieved CR after induction chemotherapy.
To examine whether miRNA-181a-3p was decreased in patients who achieved
CR after induction chemotherapy, we detected its expression from blood
specimens in 10 patients. On day 28 after induction chemotherapy,
miRNA-181a-3p expression was significantly decreased in 80% of patients
(P<0.001, Figure 2). Moreover, on day 28 after induction chemotherapy, miR-181a-3p was expressed at higher levels compared with healthy controls (P<0.05, Figure 2A).
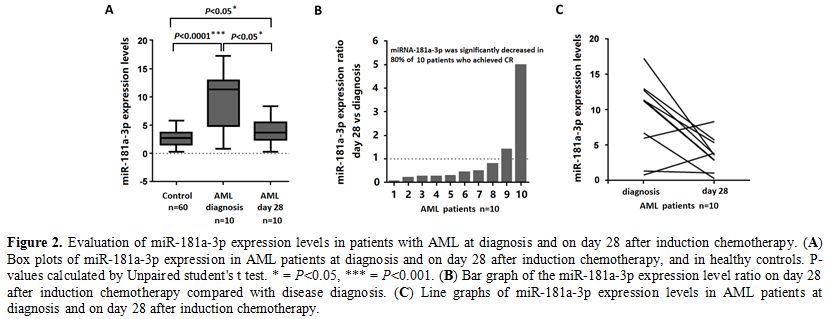 |
Figure 2. Evaluation of
miR-181a-3p expression levels in patients with AML at diagnosis and on
day 28 after induction chemotherapy. (A)
Box plots of miR-181a-3p expression in AML patients at diagnosis and on
day 28 after induction chemotherapy, and in healthy controls. P-values
calculated by Unpaired student's t test. * = P<0.05, *** =
P<0.001. (B) Bar graph of the miR-181a-3p expression level ratio on day 28 after induction chemotherapy compared with disease diagnosis. (C) Line graphs of miR-181a-3p expression levels in AML patients at diagnosis and on day 28 after induction chemotherapy. |
Association of miR181a-3p expression with AML patient outcome.
A total of 119 adult patients with newly diagnosed AML were recruited,
8 patients died within 30 days after chemotherapy, finally 111 patients
were included for statistical analysis. A total of 81 patients achieved
complete remission (CR), whereas 30 patients failed to. To graphically
display the association of miR181a-3p expression with CR achievement,
we compared expression levels in patients who achieved CR (n=81) with
those who failed to achieve CR (n=30) (Fig 3A). At the time of diagnosis, miRNA-181a-3p expression level was correlated with the response to induction chemotherapy (P<0.05).
According
to miR-181a-3p expression, patients with AML were dichotomized into
high (above median expression levels) and low (below or at median
expression levels) groups. Kaplan–Meier survival curves showed that
patients with higher miR-181a-3p expression levels at diagnosis
presented with a better OS (P=0.014, Fig 3D) , but not a better DFS (P=0.062, Fig 3C),
than those with lower expression levels. In the TCGA analysis, AML
patients with higher miR-181a-3p also presented with a better OS
(P=0.008, Fig 3E).
The
Mantel–Cox test and Gehan–Breslow–Wilcoxon test were performed to
determine the relationship between miR-181a-3p expression and OS or DFS
(Fig 3B). The
Gehan–Breslow–Wilcoxon test highlighted the prognostic value of
increased miR-181a-3p expression at diagnosis both for disease relapse
(HR: 1.597; 95% CI: 0.9804-2.559; P=0.03) and death (HR: 2.062; 95% CI: 1.160-3.456; P=0.02). Thus, a higher miR-181a-3p expression at diagnosis was significantly associated with patient outcome.
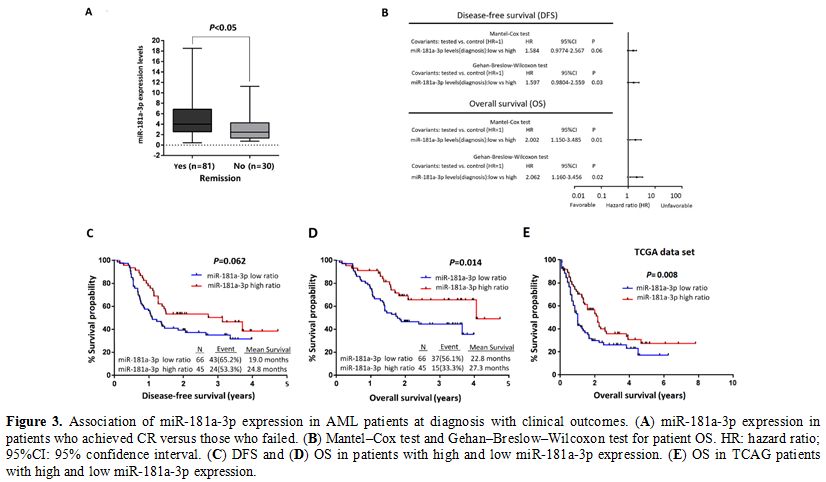 |
Figure 3. Association of miR-181a-3p expression in AML patients at diagnosis with clinical outcomes. (A) miR-181a-3p expression in patients who achieved CR versus those who failed. (B) Mantel–Cox test and Gehan–Breslow–Wilcoxon test for patient OS. HR: hazard ratio; 95%CI: 95% confidence interval. (C) DFS and (D) OS in patients with high and low miR-181a-3p expression. (E) OS in TCAG patients with high and low miR-181a-3p expression. |
Finally,
multivariate Cox analysis was performed to determine the relationship
between independent prognostic value and OS or DFS. The multivariate
Cox analysis was adjusted for patients’ age, gender, WBC count and
disease risk stratification. Multivariate analysis (Table 3) highlighted the of miR-181a-3p levels on AML diagnosis not for disease relapse (P>0.05) but for death (HR: 1.923; 95% CI: 1.035-3.574; P<0.05).
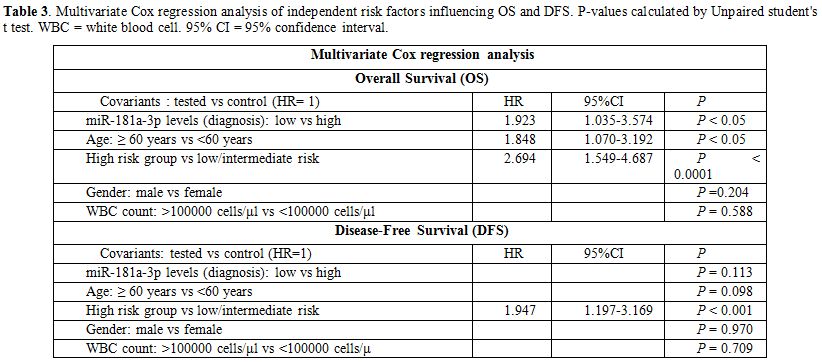 |
Table 3. Multivariate Cox
regression analysis of independent risk factors influencing OS and DFS.
P-values calculated by Unpaired student's t test. WBC = white blood
cell. 95% CI = 95% confidence interval. |
Assessment of the relationship between miR-181a-3p ectopic expression and IKBKG and NF-κB family.
Since miR-181a-3p blocks the NF-κB signaling pathway by targeting
NEMO/IKBKG in Human Umbilical Vein Endothelial Cells (HUVECs),[8]
whether IKBKG and NF-κB expressions are affected by miR-181a-3p in AML
cells needed to be determined. NF-κB family contains NF-κB1, NF-κB2,
RelA, RelB and Rel. Firstly, we investigated the relation of
miR-181a-3p ectopic expression with NEMO/IKBKG and NF-κB family in a
set of primary TCGA AML patients. NEMO/IKBKG expression was positively
correlated with expression of NF-κB family (NF-κB1, NF-κB2, RelA, RelB)
(Person correlation= 0.377, P<0.01; Person correlation= 0.598, P<0.01; Person correlation= 0.557, P<0.01; Person correlation= 0.524, P<0.01).
MiR-181a-3p expression was negatively correlated with expression of
NF-κB family (NF-κB1, NF-κB2, RelA, RelB) (Person correlation=
-0.209, P<0.05; Person correlation= -0.555, P<0.01, Person correlation= -0.19, P<0.05, Person correlation= -0.309, P<0.01) and NEMO/IKBKG (Person correlation= -0.313, P<0.01).
Then we investigated the relation of miR-181a-3p expression with
NEMO/IKBKG and NF-κB in 59 AML patients, and found that miR-181a-3p
expression was negatively correlated with the expression of NF-κB
(Person correlation= -0.2795, P=0.0321) and NEMO/IKBKG (Person correlation= -0.2613, P=0.0456). The linear correlation analysis in AML samples were showed in Table 4..
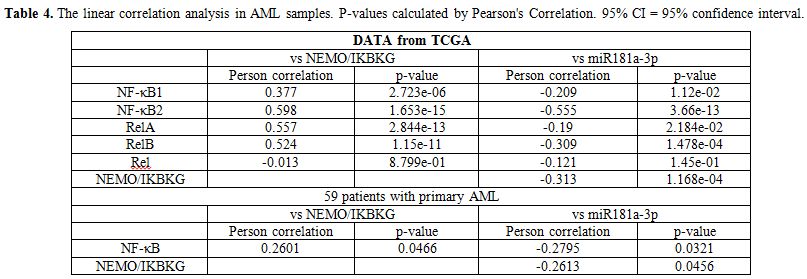 |
Table 4. The linear
correlation analysis in AML samples. P-values calculated by Pearson's
Correlation. 95% CI = 95% confidence interval. |
Discussion
In
our study, we evaluated the clinical diagnostic value and role of
miRNA-181a-3p in predicting prognosis and outcomes. We showed that
miRNA-181a-3p expression was significantly increased in AML patients
compared with controls, while ROC analyses verified the ability of
miR-181a-3p to distinguish AML from control blood samples. The
follow-up of 10 patients revealed a significant down-regulation of
miR-181a-3p expression on day 28 of induction chemotherapy, while
higher miR-181a-3p expression at diagnosis was correlated with
favorable prognosis. Finally, we found that miR-181a-3p expression was
negatively correlated with the expression of NEMO/IKBKG. Our findings
should be confirmed using a larger sample size.
In recent years,
novel high-throughput sequencing techniques have significantly advanced
our understanding of the molecular pathogenesis of AML.[10-11]
Several recurrent mutations in genes encoding epigenetic modifiers have
been identified that affect not only disease phenotype but also a
response to therapy.[12-13] MiRNAs play an important
role in the pathogenesis and development of AML, and their abnormal
expression is associated with specific cytogenetic subsets or mutations
of AML, suggesting that they could be used as independent biomarkers
for determining the outcomes of AML patients.[4,14-15]
MiR-181a
belongs to the miR-181 family, its role in tumors is still
controversial, and it may function as a tumor promoter or suppressor
depending on tumor type.[16] In hematologic
malignancies, miR-181a functions as a tumor suppressor in cellular
division and differentiation. AML patients with higher miR-181
expression at diagnosis have a better prognosis than those with lower
miR-181 expression, miR-181 may be a diagnostic biomarker and predictor
of prognosis in AML patients.[17-19]
Precursor
miR-181a can be processed into two mature strands: miR-181a-3p and
miR-181a-5p. MiR-181a-3p is highly expressed in RPMI8226 cell-derived
extracellular vesicles (R-EVs) and regulates cell proliferation.[20]
Mir-181a-3p blocks the NF-κB signaling pathway by targeting
NEMO/IKBKG in Human Umbilical Vein Endothelial Cells (HUVECs),
and miR-181a-3p mimics treatment prevents myeloid cell recruitment and
decreased the expression of TNF-α in apoE −/− mice.8 NF-κB is an
important transcription factor, which plays a crucial cancer-promoting
role in Acute myeloid leukemia (AML).[21-22] It has
been known that chromosomal translocations or gene mutations leading to
the increase in NF-κB activity. NEMO/IKBKG acts as a crucial
antiapoptotic transcription factor, which is crucial for the activation
of NF-κB.[23] NF-κB family contains RelA, RelB,
NF-κB1, NF-κB2 and Rel. We found that miR-181a-3p expression was
negatively correlated with the expression of NF-κB family (NF-κB1,
NF-κB2, RelA, RelB) and NEMO/IKBKG in TCGA samples. In our study,
miR-181a-3p expression was negatively correlated with the expression of
NF-κB and NEMO/IKBKG in 59 AML patients. Maybe mir-181a-3p affects AML
cell proliferation and apoptosis by targeting NEMO/IKBKG. We should
make more efforts to test this hypothesis.
In summary, we reported
a clinical role for miR-181a-3p in AML patients for the first time.
MiRNA-181a-3p expression was shown to have value in discriminating AML
patients from healthy controls, and to correlate with the response to
induction chemotherapy. Patients with higher miR-181a-3p expression
levels at diagnosis demonstrated an improved OS. Therefore, the
miR-181a-3p expression may be an independent prognostic biomarker for
AML patient outcomes.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 81572065)
References
- Coombs CC, Tallman MS, Levine RL: Molecular therapy
for acute myeloid leukaemia. Nature Reviews Clinical Oncology 2016,
13(5):305-318. https://doi.org/10.1038/nrclinonc.2015.210 PMid:26620272 PMCid:PMC5525060
- Marcucci
G, Mrozek K, Radmacher MD, Garzon R, Bloomfield CD: The prognostic and
functional role of microRNAs in acute myeloid leukemia. Blood 2011,
117(4):1121-1129. doi:10.1182/blood-2010-09-191312. https://doi.org/10.1182/blood-2010-09-191312 PMid:21045193 PMCid:PMC3056468
- Wallace
JA, O'Connell RM: MicroRNAs and acute myeloid leukemia: therapeutic
implications and emerging concepts. Blood 2017, 130(11):1290-1301. https://doi.org/10.1182/blood-2016-10-697698 PMid:28751524 PMCid:PMC5600138
- Diaz-Beya
M, Brunet S, Nomdedeu J, Tejero R, Diaz T, Pratcorona M, Tormo M,
Ribera JM, Escoda L, Duarte R et al: MicroRNA expression at diagnosis
adds relevant prognostic information to molecular categorization in
patients with intermediate-risk cytogenetic acute myeloid leukemia.
Leukemia 2014, 28(4):804-812. https://doi.org/10.1038/leu.2013.281 PMid:24072101
- Chuang
MK, Chiu YC, Chou WC, Hou HA, Chuang EY, Tien HF: A 3-microRNA scoring
system for prognostication in de novo acute myeloid leukemia patients.
Leukemia 2015, 29(5):1051-1059. https://doi.org/10.1038/leu.2014.333 PMid:25428263
- Yang
Z, Wan X, Gu Z, Zhang H, Yang X, He L, Miao R, Zhong Y, Zhao H:
Evolution of the mir-181 microRNA family. Computers in Biology and
Medicine 2014, 52:82-87. https://doi.org/10.1016/j.compbiomed.2014.06.004 PMid:25016292
- Su
R, Lin HS, Zhang XH, Yin XL, Ning HM, Liu B, Zhai PF, Gong JN, Shen C,
Song L et al: MiR-181 family: regulators of myeloid differentiation and
acute myeloid leukemia as well as potential therapeutic targets.
Oncogene 2015, 34(25):3226-3239. https://doi.org/10.1038/onc.2014.274 PMid:25174404
- Su
Y, Yuan J, Zhang F, Lei Q, Zhang T, Li K, Guo J, Hong Y, Bu G, Lv X et
al: MicroRNA-181a-5p and microRNA-181a-3p cooperatively restrict
vascular inflammation and atherosclerosis. Cell Death & Disease
2019, 10(5):365. https://doi.org/10.1038/s41419-019-1599-9 PMid:31064980 PMCid:PMC6504957
- Sun
XX, Zhang SS, Dai CY, Peng J, Pan Q, Xu LF, Ma XL: LukS-PV-Regulated
MicroRNA-125a-3p Promotes THP-1 Macrophages Differentiation and
Apoptosis by Down-Regulating NF1 and Bcl-2. Cellular physiology and
biochemistry. International Journal of Experimental Cellular
Physiology, Biochemistry, and Pharmacology 2017, 44(3):1093-1105. https://doi.org/10.1159/000485415 PMid:29179212
- Bullinger
L, Dohner K, Dohner H: Genomics of Acute Myeloid Leukemia Diagnosis and
Pathways. Journal of clinical oncology : official journal of the
American Society of Clinical Oncology 2017, 35(9):934-946. https://doi.org/10.1200/JCO.2016.71.2208 PMid:28297624
- Ferrando AA, Lopez-Otin C: Clonal evolution in leukemia. Nature Medicine 2017, 23(10):1135-1145. https://doi.org/10.1038/nm.4410 PMid:28985206
- Short NJ, Rytting ME, Cortes JE: Acute myeloid leukaemia. Lancet (London, England) 2018, 392(10147):593-606. https://doi.org/10.1016/S0140-6736(18)31041-9
- Medinger M, Passweg JR: Acute myeloid leukaemia genomics. British Journal of Haematology 2017, 179(4):530-542. https://doi.org/10.1111/bjh.14823 PMid:28653397
- de
Leeuw DC, Verhagen HJ, Denkers F, Kavelaars FG, Valk PJ, Schuurhuis GJ,
Ossenkoppele GJ, Smit L: MicroRNA-551b is highly expressed in
hematopoietic stem cells and a biomarker for relapse and poor prognosis
in acute myeloid leukemia. Leukemia 2016, 30(3):742-746. https://doi.org/10.1038/leu.2015.160 PMid:26108690
- Marcucci
G, Radmacher MD, Mrozek K, Bloomfield CD: MicroRNA expression in acute
myeloid leukemia. Current Hematologic Malignancy Reports 2009,
4(2):83-88. https://doi.org/10.1007/s11899-009-0012-7 PMid:20425419
- Roth E, Cao J: MiR-181 suppresses metastasis via MMP-14. Aging 2015, 7(10):740-741. https://doi.org/10.18632/aging.100824 PMid:26527609 PMCid:PMC4637197
- Schwind
S, Maharry K, Radmacher MD, Mrozek K, Holland KB, Margeson D, Whitman
SP, Hickey C, Becker H, Metzeler KH et al: Prognostic significance of
expression of a single microRNA, miR-181a, in cytogenetically normal
acute myeloid leukemia: a Cancer and Leukemia Group B study. Journal of
clinical oncology. Official Journal of the American Society of Clinical
Oncology 2010, 28(36):5257-5264. https://doi.org/10.1200/JCO.2010.29.2953 PMid:21079133 PMCid:PMC3018359
- Butrym
A, Rybka J, Baczynska D, Poreba R, Mazur G, Kuliczkowski K: Expression
of microRNA-181 determines response to treatment with azacitidine and
predicts survival in elderly patients with acute myeloid leukaemia.
Oncology Letters 2016, 12(4):2296-2300. https://doi.org/10.3892/ol.2016.4970 PMid:27698792 PMCid:PMC5038519
- Weng
H, Lal K, Yang FF, Chen J: The pathological role and prognostic impact
of miR-181 in acute myeloid leukemia. Cancer Genetics 2015,
208(5):225-229. https://doi.org/10.1016/j.cancergen.2014.12.006 PMid:25686674 PMCid:PMC4466067
- Zhang
L, Lei Q, Wang H, Xu C, Liu T, Kong F, Yang C, Yan G, Sun L, Zhao A et
al: Tumor-derived extracellular vesicles inhibit osteogenesis and
exacerbate myeloma bone disease. Theranostics 2019, 9(1):196-209. https://doi.org/10.7150/thno.27550 PMid:30662562 PMCid:PMC6332790
- Kagoya
Y, Yoshimi A, Kataoka K, Nakagawa M, Kumano K, Arai S, Kobayashi H,
Saito T, Iwakura Y, Kurokawa M: Positive feedback between NF-kappaB and
TNF-alpha promotes leukemia-initiating cell capacity. The Journal of
Clinical Investigation 2014, 124(2):528-542. https://doi.org/10.1172/JCI68101 PMid:24382349 PMCid:PMC3904603
- Bosman
MC, Schuringa JJ, Vellenga E: Constitutive NF-kappaB activation in AML:
Causes and treatment strategies. Critical reviews in
oncology/hematology 2016, 98:35-44. https://doi.org/10.1016/j.critrevonc.2015.10.001 PMid:26490297
- Brahler
S, Ising C, Barrera Aranda B, Hohne M, Schermer B, Benzing T,
Brinkkoetter PT: The NF-kappaB essential modulator (NEMO) controls
podocyte cytoskeletal dynamics independently of NF-kappaB. American
Journal of Physiology Renal Physiology 2015, 309(7):F617-626. https://doi.org/10.1152/ajprenal.00059.2015 PMid:26268269
[TOP]






