Hongbo Hu1, Ying Cheng2, Qiaoying Peng3 and Kun Chen4..
1 Department of Laboratory, Maternal and Child Health Hospital of Hubei Province, China.
2 Department of Pediatrics, Maternal and Child Health Hospital of Hubei Province, China.
3 Department of Neonatology, Maternal and Child Health Hospital of Hubei Province, China.
4 Department of Laboratory, Wuhan Ninth Hospital, China.
Correspondence to: Kun Chen, Department of Laboratory, Wuhan Ninth Hospital, No.
20, Jilin Street, Qingshan District, Wuhan 430081, China. Tel:
86-027-68865331. E-mail:
chenkun430922@163.com
Published: September 1, 2020
Received: May 25, 2020
Accepted: August 4, 2020
Mediterr J Hematol Infect Dis 2020, 12(1): e2020057 DOI
10.4084/MJHID.2020.057
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
To the editor,
Genotyping
of CMV has mainly focused on gB, gN, and gH, which play a role in virus
entry and may influence the infectivity or pathogenicity of CMV.[1,2]
It has been hypothesized that genetic variation among CMV strains may
underlie strain-specific clinical manifestations. Our previous research
revealed that there might be a potential association between the
genotypes of CMV and neonatal thrombocytopenia, and the detection of
some specific genotypes might be indicative of severe manifestations in
infants with CMV infection.[3,4] However, the study
design and the criteria to define the study population (congenital and
non-congenital cases) and the setting of the control group
(CMV-associated thrombocytopenia and non-thrombocytopenic cases) were
not clearly established. For this reason, we included patients
classified on more unambiguous criteria, and the clinical data
collected were complete and thoroughly detailed, which allows us to
assess the association between genotypes and the outcome in the
non-congenital population.
Methods
Definition.
Symptomatic perinatal infection is defined as an infant presenting CMV
associated symptoms and positive CMV detection in 3-12 weeks after
birth. Symptomatic postnatal infection is referred to as an infant
presenting CMV associated symptoms and positive CMV detection after 12
weeks of birth.[5] Altogether, in the present study,
both of them referred to as CMV symptomatic postnatal infection.
Moderately to severely symptomatic CMV disease is defined as multiple
manifestations attributable to CMV infection. Mildly symptomatic CMV
disease is characterized as by one or two isolated features of CMV
infection that are mild and transient (e.g., mild hepatomegaly or a
single measurement of low platelet count or raised levels of alanine
aminotransferase).[6]
Patients.
Thirty immunocompetent patients (median, two months; range, 25 days–11
months) with CMV-associated thrombocytopenia were analyzed, including
18 perinatal infections and 12 postnatal infections. Of these 30
patients, 20 were diagnosed with moderately to severely symptomatic CMV
disease, and 10 were diagnosed with mildly symptomatic CMV disease. The
clinical records of the 30 postnatally infected infants are summarized
in Table 1. A group of 40
non-thrombocytopenic individuals, including 20 asymptomatic infants
(median, two months; range, 25 days–10 months) and 20 patients (median,
two months; range, 29 days–11 months) in CMV infections involving organ
systems other than the hematopoietic system from the same period was
also included in the study. Among 20 non-thrombocytopenic patients,
respiratory symptoms including upper respiratory tract infection
(20.0%, 4/20), bronchitis (25.0%, 5/20), and pneumonia (30.0%, 6/20)
were the most common symptom at presentation. Other presentations were
hepatitis (10.0%, 2/20), jaundice (25.0%, 5/20), and 1 case (5.0%,
1/20) had cholestasis. The baseline characteristics and clinical
manifestations in these infants have been described in Table 1 and Table 2.
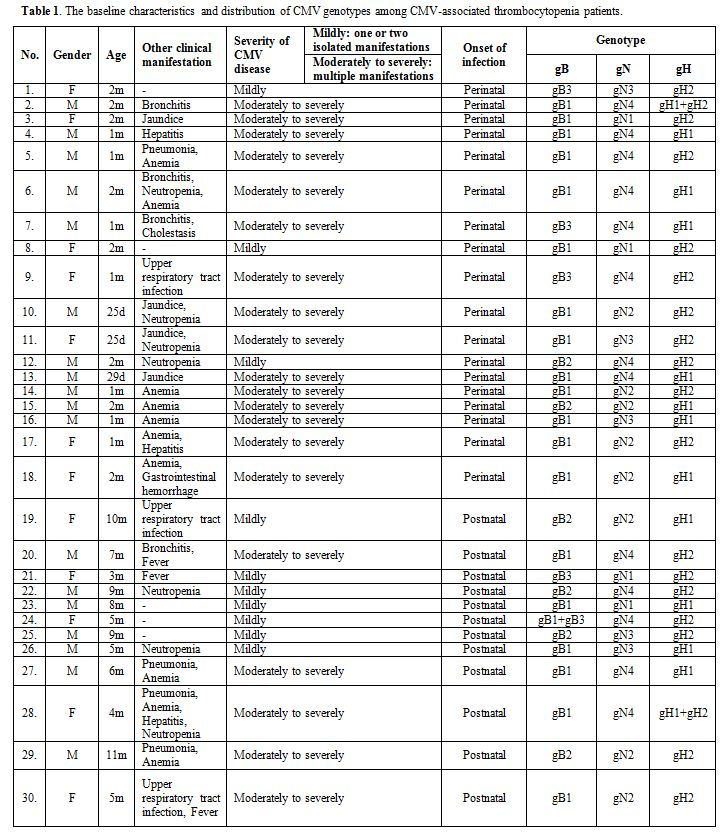 |
Table 1.
The baseline characteristics and distribution of CMV genotypes among CMV-associated thrombocytopenia patients. |
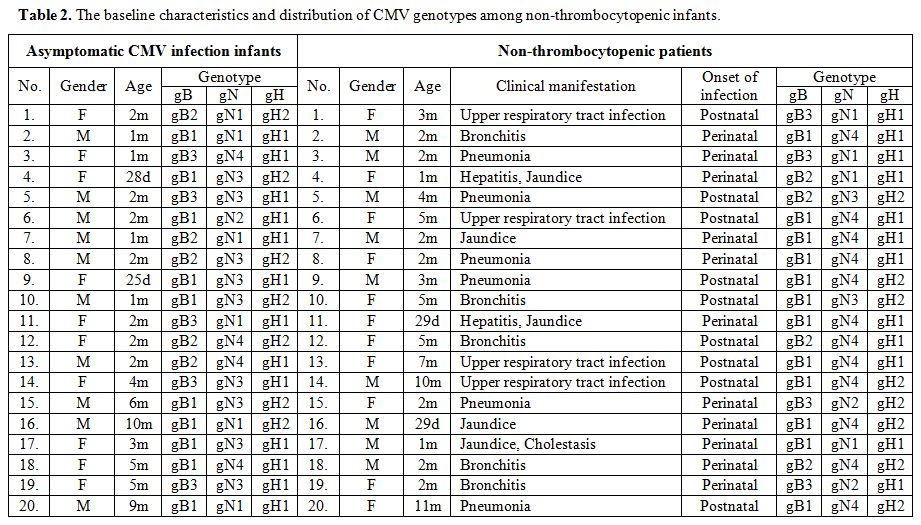 |
Table 2. The baseline characteristics and distribution of CMV genotypes among non-thrombocytopenic infants. |
Laboratory test for CMV infection.
Patients were tested for CMV infection using serological CMV tests (IgM
and IgG), viral culture, and real-time PCR for blood or urine samples.
CMV IgM and CMV IgG were tested using an ELISA kit according to the
manufacturer's instructions (DiaSorin S.p.A., Italy). For testing CMV
in urine, urine samples were collected and cultured using the shell
vial culture method (Chemicon, Temecula, CA, USA). According to the
manufacturer's instructions (Daan Gene Company of Zhongshan University,
China), fluorescence quantitative CMV-DNA kit was used to quantify of
CMV-DNA. DNA level > 103 copies/ml
indicated replication, which was considered positive in this study. CMV
gB,gN and gH genotype analysis was done by nested PCR and restriction
length polymorphism as reported.[7-9]
Statistical analyses.
Statistical analysis was conducted using the SPSS ver. 21.0 software
(SPSS, Inc., Chicago, IL, USA). Genotype distribution among postnatally
infected patients, the relationship between the gB, gN, and gH
genotypes and the severity of CMV infections were analyzed using the
chi-square test for ratio comparison. Logistic regression analysis was
used to assess the associated risk between particular genotypes and the
variables of the study. A P-value of less than 0.05 was considered to
be statistically significant.
Results
CMV Genotyping.
The distribution of gB genotypes in this present study was gB1 (63.3%,
19/30), followed by gB2 (20.0%, 6/30) and gB3 (13.3%, 4/30). We also
found 1 coinfection case (3.3%, 1/30) with 2 genotypes (gB1/gB3), no
gB4 genotype was found. Notably, significantly higher frequency of gB1
(80.0%,16/20) was found in moderately to severely CMV infection infants
compared to infants with mildly symptomatic CMV disease ( χ2= 8.132, p
= 0.043) (Figure 1).
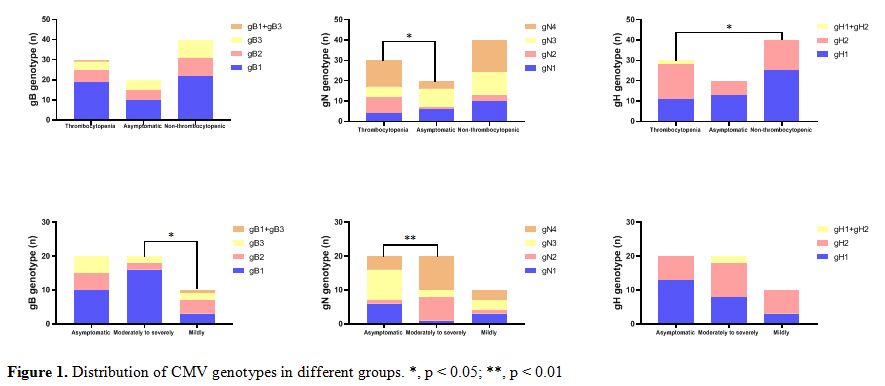 |
Figure 1. Distribution of CMV genotypes in different groups. *, p < 0.05; **, p < 0.01 |
The
overall distribution of individual genotypes in this study cohort was
as follows: gN1(13.3%,4/30), gN2 (26.7%,8/30), gN3 (16.7%,5/30) and gN4
(43.3%,13/30). Comparing distribution in 20 asymptomatic infants with
CMV infection, the gN1 (5.0%,1/20) was the less prevalent genomic
variants in moderately to severely CMV infection patients (χ2=15.097, p
= 0.002) (Figure 1).
The
gH1, gH2 and gH1/gH2 genotypes were distributed in 36.7% (11/30), 56.7%
(17/30) and 6.7% (2/30) of the patients, respectively (Figure 1).
Compared with the genotype distribution in non-thrombocytopenic
infants, a greater frequency of gH2 in CMV-associated thrombocytopenia
infants was noted with significant difference (χ2=6.269, p
= 0.044). No difference in the distribution of gH genotypes in
symptomatic and asymptomatic patients, or in moderately to severely
symptomatic CMV disease and mildly symptomatic CMV disease(Figure 1).
Genotype Association With CMV-associated thrombocytopenia and severity of CMV disease. In the logistic regression analysis, the gN2 [p = 0.043, with OR=4.598, 95%CI (1.052-20.098)] and gH2 [p
= 0.038, with OR=2.933, 95%CI (1.060-8.117)] genotypes were associated
with an elevated risk of developing thrombocytopenia. Besides, gB1 [p
= 0.022, with OR=9.820, 95%CI (1.400-68.888)] represented the most
virulent genotypes and was associated with severe manifestations in
CMV-associated thrombocytopenia infants. Conversely, the gN1 [p = 0.044, with OR=0.061, 95%CI (0.004-0.930)] genotype was associated with a reduced risk of severely symptomatic CMV disease.
Discussion
The
gB of CMV likely plays a crucial role in viral entry into cells, the
transmission of the virus from cell to cell, and the fusion of infected
cells. It has been reported that the gB genotypes vary in their ability
to stimulate cell-mediated or cytotoxic immune response.[10,11]
Therefore, variations in gB are likely to have significant effects on
the pathogenesis of CMV disease and the spectrum of host cells infected
by the virus. Our previous studies also confirmed that the gB1 genotype
had more virulence in infants with symptomatic CMV disease.[3,4]
But
interestingly, in asymptomatic infected infants, gB1 was also the
dominant genotype, and its genotype distribution was not significantly
different from that of CMV-associated thrombocytopenia infants.
Consequently,we speculate that CMV gB1 strains may elicit a severe
immunopathological response that in some infants can control the
symptoms of CMV and, in others, lead to CMV-associated thrombocytopenia
with organ damage and disease manifestations. However, the virulence of
gB1 in asymptomatic infants is negligible in relationship with a
difference in the individual immune status.
The CMV strain with
gN1 genotype may represent a less virulent virus phenotype, especially
considering that the variation is a typical AD169-like glycoprotein,
which is far away from CMV clinical isolates in immunology.[12-14]
In our study, among CMV-associated thrombocytopenia infants (20 cases)
who were classified as having moderately to severely symptomatic CMV
disease, 17 had gN4 or gN2 genotypes and only one had a gN1 genotype,
supporting the idea that gN1 genotype may be less virulent. In
addition, compared with the genotype distribution in asymptomatic and
non-thrombocytopenic infants in present study, thrombocytopenia
occurred more frequently in infants infected with the CMV gN2 genotype,
although the proportion of this genotype was less than that of gN4 in
CMV-associated thrombocytopenia infants. The gN2 genotype was detected
in 26.7% (8/30) of infants with CMV-associated thrombocytopenia and was
associated with at least a 4-fold increased risk of developing
thrombocytopenia. Our study is the first to demonstrate that a gN
variant might be associated with a risk of CMV-associated
thrombocytopenia in infants infected postnatally.
As we reported
earlier, the gH2 genotype was associated with at least a 7-fold
increased risk of developing CMV-associated thrombocytopenia among
infants with congenital and perinatal infections.[4] After including postnatal infection and non-thrombocytopenic cases into the analysis, similar conclusions were reached.
Based
on these cases, several general points can be highlighted. First, in
regression analysis, the difference in the setting of the
non-thrombocytopenic control group, which includes asymptomatic and
symptomatic infants, may cause a discrepancy in results. Increasing the
sample size and choosing an appropriate scale setting may reduce this
discrepancy. Second, a specific cytomegalovirus genotype may show
strong virulence in some CMV- related diseases, while in other
CMV-related diseases or asymptomatic infants, it may not show
corresponding characteristics of virulence. Finally, in addition to CMV
gB, gN, and gH, CMV glycoprotein also includes gO, gM and gL. Six
glycoproteins are essential for fibroblasts to enter CMV, and form
glycoprotein complexes, gCI (gB), gCII (gM / gN), gcIII (gH / gL / gO)
on the virus membrane.[15] In the study of a CMV-
related disease, it is more reasonable to include all essential CMV
glycoprotein genotypes into the analysis.
References
- Ross SA , Pati P, Jensen TL, et al. Cytomegalovirus
genetic diversity following primary infection. J Infect Dis.
2020;221(5):715-720. https://doi.org/10.1093/infdis/jiz507 PMid:31593588
- Nahar
S, Hokama A, Iraha A, et al. distribution of cytomegalovirus genotypes
among ulcerative colitis patients in Okinawa, Japan. Intest Res. 2018
;16(1):90-98. https://doi.org/10.5217/ir.2018.16.1.90 PMid:29422803 PMCid:PMC5797277
- Hu
H, Cheng Y, Peng Q, Chen K. Clinical Features, Treatment Courses, and
Distribution of Cytomegalovirus Genotypes among Thrombocytopenia
Patients Aged Younger than 12 Months [published online ahead of print,
2020 Jun 11]. Am J Perinatol. 2020;10. https://doi.org/10.1055/s-0040-1713001
- Hu
H, Peng W, Peng Q, Cheng Y. Cytomegalovirus Genotype Distribution among
Congenital and Perinatal Infected Patients with CMV-Associated
thrombocytopenia [published online ahead of print, 2020 Jun 1]. Fetal
Pediatr Pathol. 2020;1-10. https://doi.org/10.1080/15513815.2020.1765916 PMid:32479132
- Shen
Z, Shang SQ, Zou CC, Zheng JY, Yu ZS. The detection and clinical
features of human cytomegalovirus infection in infants. Fetal Pediatr
Pathol. 2010;29(6):393-400. https://doi.org/10.3109/15513815.2010.494705 PMid:21043563
- Rawlinson
WD, Boppana SB, Fowler KB, et al. Congenital cytomegalovirus infection
in pregnancy and the neonate: consensus recommendations for prevention,
diagnosis, and therapy. Lancet Infect Dis. 2017;17(6): e177-e188. https://doi.org/10.1016/S1473-3099(17)30143-3
- Yu
ZS, Zou CC, Zheng JY, Zhao ZY. Cytomegalovirus gB genotype and clinical
features in Chinese infants with congenital infections. Intervirology.
2006;49(5):281-285. https://doi.org/10.1159/000093458 PMid:16714857
- Guo
S, Yu MM, Li G, Zhou H, Fang F, Shu SN. Studies on genotype of human
cytomegalovirus glycoprotein H from infantile clinical isolates.
Zhonghua Er Ke Za Zhi. 2013;51(4):260-264 (in Chinese).
- Chen
HP, Lin JC, Yang SP, et al. The type-2 variant of human cytomegalovirus
glycoprotein N(gN-2) is not the rarest in the Chinese population of
Taiwan: influence of primer design. J Virol
Methods.2008;151(1):161-164. https://doi.org/10.1016/j.jviromet.2008.03.018 PMid:18499272
- Saccoccio
FM , Jenks JA , Itell HL, et al. Humoral immune correlates for
prevention of postnatal cytomegalovirus acquisition. J Infect Dis.
2019;220(5):772-780. https://doi.org/10.1093/infdis/jiz192 PMid:31107951 PMCid:PMC6667799
- Lee
S, Doualeh M , Affandi JS , Makwana N , Irish A , Price P. Functional
and clinical consequences of changes to natural killer cell phenotypes
driven by chronic cytomegalovirus infections. J Med Virol.
2019;91(6):1120-1127. https://doi.org/10.1002/jmv.25401 PMid:30636352
- Paradowska
E, Jabłońska A, Studzińska M, et al. Distribution of cytomegalovirus gN
variants and associated clinical sequelae in infants. J Clin Virol.
2013;58(1):271-275. https://doi.org/10.1016/j.jcv.2013.05.024 PMid:23806667
- Pignatelli
S , Rossini G, Dal Monte P, Gatto MR, Landini MP. Human cytomegalovirus
glycoprotein N genotypes in AIDS patients. AIDS. 2003;
28;17(5):761-763. https://doi.org/10.1097/00002030-200303280-00018 PMid:12646803
- Mujtaba
G, Khurshid A, Sharif S, et al. distribution of cytomegalovirus among
neonates born to infected mothers in Islamabad, Pakistan. PLoS One.
2016 ;11(7): e0156049. https://doi.org/10.1371/journal.pone.0156049 PMid:27367049 PMCid:PMC4930188
- Coleman
S, Hornig J, Maddux S, et al. Viral glycoprotein complex formation,
essential function and immunogenicity in the guinea pig model for
cytomegalovirus. PLoS One. 2015;12;10(8): e0135567. https://doi.org/10.1371/journal.pone.0135567 PMid:26267274 PMCid:PMC4534421
[TOP]


