Matteo Molica1, Elisabetta Abruzzese1,2 and Massimo Breccia3.
1 Haematology Unit, S. Eugenio Hospital, Rome, Italy
2 Tor Vergata University Hospital, Department of Hematology, Rome, Italy
3 Hematology, Department of Cellular Biotechnologies and Hematology, Policlinico Umberto 1, Sapienza University, Rome, Italy
Correspondence to: Matteo Molica. Haematology Unit, S. Eugenio Hospital, Rome, Italy. E-mail:
molica@bce.uniroma1.it
Published: September 1, 2020
Received: June 2, 2020
Accepted: August 10, 2020
Mediterr J Hematol Infect Dis 2020, 12(1): e2020062 DOI
10.4084/MJHID.2020.062
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Chronic myeloid leukemia (CML) is characterized by the presence of the BCR-ABL1 fusion gene. In more than 95% of CML patients, the typical BCR-ABL1
transcript subtypes are e13a2 (b2a2), e14a2 (b3a2), or the simultaneous
expression of both. Other less frequent transcript subtypes, such as
e1a2, e2a2, e6a2, e19a2, e1a3, e13a3, and e14a3, have been sporadically
reported. The main purpose of this review is to assess the possible
impact of different transcripts on the response rate to tyrosine kinase
inhibitors (TKIs), the achievement of stable deep molecular responses
(s-DMR), the potential maintenance of treatment-free remission (TFR),
and long-term outcome of CML patients treated with TKIs. According to
the majority of published studies, patients with e13a2 transcript
treated with imatinib have lower and slower cytogenetic and molecular
responses than those with e14a2 transcript. They should be considered a
high-risk group that would most benefit from frontline treatment with
second-generation TKIs (2GTIKIs). Although few studies have been
published, similar significant differences in response rates to 2GTKIs
have been not reported. The e14a2 transcript seems to be a favorable
prognostic factor for obtaining s-DMR, irrespective of the TKI
received, and is also associated with a very high rate of TFR
maintenance. Indeed, patients with e13a2 transcript achieve a lower
rate of s-DMR and experience a higher probability of TFR failure.
According to most reported data in the literature, the type of
transcript does not seem to affect long-term outcomes of CML patients
treated with TKIs. In TFR, the e14a2 transcript appears to be related
to favorable responses. 2GTKIs as frontline therapy might be a
convenient approach in patients with e13a2 transcript to achieve
optimal long-term outcomes.
|
Introduction
Chronic
myeloid leukemia (CML) is a hematological malignancy that has an
estimated incidence of 1-2 cases per 100,000 adults and accounts for
nearly 15% of new leukemia diagnoses in adults. The prevalence of CML
in the US was estimated at approximately 80,000-100,000+ in 2017.[1]
Pathognomonic CML is a cytogenetic aberration well-known as the
Philadelphia (Ph) chromosome, which represents the first chromosome
alteration associated with a specific human malignancy.[2] The Philadelphia chromosome derives from a reciprocal translocation involving the 3′ region of the proto-oncogene c-ABL (9q34) and the 5′ region of the breakpoint cluster region (BCR) gene on chromosome 22q11 (t(9;22)(q34;11). This balanced translocation determines the production of the BCR-ABL1 oncogene, which encodes a protein with constitutive tyrosine kinase activity that promotes leukemogenesis.[3,4] The major breakpoint cluster region (M-BCR) consists of 5 exons called e12 to e16 (formerly b1-b5) located within the central region of the BCR
gene, and more than 95% of CML patients have a breakpoint in this
specific region. Two major breakpoints are identified, one after the 13th exon producing b2a2 (e13a2) fusion and the other after the 14th exon consisting of b3a2 (e14a2) fusion.[5,6]
These fusion mRNAs encode two 210-kDa tyrosine kinase proteins
(p210BCR-ABL). Approximately 5‐10% of CML patients may co-express both
e13a2 and e14a2 transcripts. These proteins act as tyrosine kinases,
have masses of 210-kDa, and differ by 25 amino acids coded by the b3
exon and an amino acid substitution (Glu903Asp).[7]
The difference between the two proteins was found in the secondary
structural elements, specifically, in five α-helices and nine β-strands
related to differences in the SH1, SH2, SH3, and DNA-binding domains.
These variations may explain the distinct activities exerted by the two
isoforms in mediating signal transduction during the evolution of the
disease.[8,9] The p210 protein is also detected in
nearly 40% of adults and 10% of children with t(9;22)-positive
precursor B-lymphoblastic leukemia (B-ALL).[10] In
approximately 60-80% of patients with Ph-positive acute lymphoblastic
leukemia (ALL) and rare cases of CML, the breakpoint occurs in the
minor-BCR (m-BCR) region, thereby resulting in the shorter isotype p190 BCR-ABL1 encoded from the e1a2 type mRNA.[11]
In CML, the e1a2 transcripts may be co-expressed with e13a2
(b2a2)/e14a2 (b3a2) and CML presenting only the e1a2 transcript is
uncommon (approximately 1% of all CML) and shows an inferior outcome to
treatment with TKIs.[12,13] An extreme 3' breakpoint seldom arises after exon 19 (e19) of BCR in the designated μ-BCR region and produces the larger (p230) fusion protein. P230BCR-ABL has been associated with a rare disease known as CML with neutrophil's maturation.[14] Several atypical BCR-ABL1
transcripts (e1a3, e13a3, e14a3, e19a3, e6a2, and e8a2, which account
for less than 1% of CML cases) deriving from chromosomal breakpoints
outside the ABL intron 1 or BCR intron 1, 13, or 14 have been described.[15] The most frequent breakpoint regions in the c-ABL
gene are 5′ of the second exon resulting in a2 junctions. Other
breakpoint regions detected between the second and the third exon have
been observed, determining a3 junctions.[16]
The BCR-ABL1 gene
can be studied by several molecular techniques (fluorescence in situ
hybridization [FISH], Southern blotting, and reverse-transcription
polymerase chain reaction [RT-PCR]). RT-PCR is the most common method
used for detecting BCR/ABL1
transcript type due to its simplicity, rapidity, and sensitivity.
However, recent new molecular techniques have been developed to detect
all kinds of transcripts using a more rapid and appropriate approach.
The multiplex PCR technique applying primers coupled to distinct
fluorochromes and the optical system of a sequencer can simultaneously
detect either the transcript (fluorescence) or the class of junction
that it holds (size).[17] This technique accurately identifies the transcript at diagnosis and allows follow-up at the molecular level.[17,18]
RT-qualitative/quantitative PCR is useful to identify the typical
transcripts (e13a2 or e14a2) at baseline and to monitor their
quantitative fluctuations during the treatment. Atypical transcripts
may yield a false negative PCR using routine primer/probe sets in
qualitative or quantitative reverse transcriptase PCR protocols. If not
tested at diagnosis, a false impression may be given that a patient is
in complete molecular response after TKI treatment. Therefore,
cytogenetics should be done in patients with atypical BCR-ABL1
transcripts that cannot be measured by RT-quantitative PCR. FISH
monitoring may also need in patients with atypical transcripts.
Several studies investigated the impact of BCR‐ABL1
transcript types on CML patients receiving TKIs; the patients'
characteristics at baseline, TKI response, and long‐term outcomes
provided different results by transcript type. In treatment-free
remission (TFR), it is essential to evaluate whether the transcript
type may identify a group of patients who more likely may achieve an
s-DMR and may have a high probability of maintaining DMRs during drug
discontinuation. In this review, we evaluated the impact of different BCR‐ABL1 transcripts on responses, long-term outcomes, and TFR rates in CML patients in TKIs.
Relationship Between Transcript Type and Outcome in pre-TKIs
In
conventional chemotherapy and interferon (IFNα), different studies have
evaluated whether the type of transcript identified at baseline may
affect the outcome of CML patients;[19,20,21]
overall, none of these studies found a significant and robust influence
of transcript type on response and clinical outcome in this setting.
However, in pre-TKI, the breakpoint in the 3' portion of the BCR region
was associated with more aggressive disease and faster transformation
in blast crisis.
In 1989, Mills et al. mapped the breakpoint
within the BCR in peripheral blood leukocyte-derived DNA from 22 CML
patients and first studied whether there was a correlation between the
site of breakpoint and outcomes. No associations between the breakpoint
site and the disease's clinical phase emerged. Still, a notable
relationship between the breakpoint site, length of time elapsed from
the presentation, and occurrence of acute phase was reported. Indeed,
the median time of chronic phase duration in patients harboring a 3'
breakpoint was 52 weeks, while that in patients with a 5' breakpoint
was 203 weeks and the rate of progression to blast crisis was
significantly different between the two groups (p<0.02).[19]
The authors concluded that patients with a 3' breakpoint had worse
outcomes, showing a four-fold more rapid transition to blast crisis
then patients with a 5' breakpoint.
Later, an English group
analyzed the correlation between mRNA transcripts and clinical
characteristics, cytogenetic response, duration of chronic phase, and
outcomes in a large cohort of 216 CML patients treated with IFNα. No
differences were found between clinical characteristics (hemoglobin
concentration, white cell and platelet count, basophil numbers, blast
cell numbers, and spleen and liver size) of patients with e13a2 and
e14a2 breakpoints except for a Sokal risk group, which was inferior
among those with e13a2 transcripts (p=0.04). No significant differences
were also observed in terms of the duration of the chronic phase and
outcomes in patients with the e13a2 and e14a2 transcripts. Five-year
survival was 52% and 54% (p=0.95) for e13a2 and e14a2, respectively.[21]
An
Italian group reported the transcript type impact on outcomes in 146
CML patients who were enrolled in a prospective study that provided
IFNα treatment for at least one year. A trend in favor of e14a2 cases
was observed or in cytogenetic response after 1 year of IFNα treatment
(39% in the e14a2 group vs 24% in the e13a2 group) in 5-year survival
rates (71% in e14a2 patients vs 57% in e13a2 patients) (Table 1).[22]
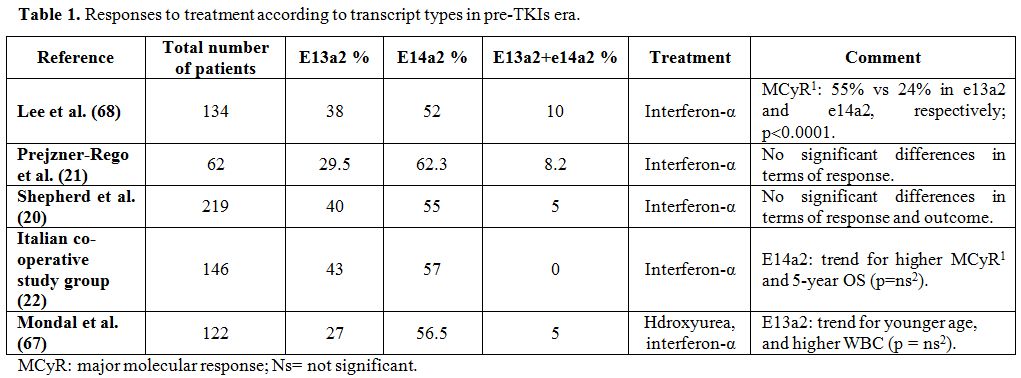 |
Table 1. Responses to treatment according to transcript types in pre-TKIs era. |
In
IFNα, although e13a2 transcript was associated with an unfavorable
trend in outcomes and treatment responses, no data were sufficient to
define the type of transcript as an independent prognostic factor.
Type of Transcript and Response to Imatinib
In the currently available literature, several studies[23,24,25,26,27,28,29,30,79] analyzed whether the two transcripts (e14a2 and e13a2) have different or similar responses to imatinib treatment.
A
German CML study group analyzed a large cohort of 1,105 newly diagnosed
imatinib-treated patients by transcript type at baseline (e13a2, n=451;
e14a2, n=496; and e13a2+e14a2, n=158). Patients expressing e14a2
transcript showed a better cumulative incidence (CI) of major molecular
response (MMR) (p=0.002) than those with e13a2 transcripts, while
patients co-expressing e13a2 and e14a2 transcripts did not differ from
the other two groups (p=ns). There was also a significant difference in
median time to MMR comparing patients harboring e13a2 and e14a2
transcripts, respectively (18.4 vs. 14.2 months). The CI of MR4.0 and
the median time to MR4 (55.2 vs. 32.4 months) were significantly better
in the e14a2 group (p<0.001). Patients co-expressing e13a2 and e14a2
transcript differed from e14a2 (p=0.004) but not from e13a2 in terms of
MR4 achievement. Patients were also evaluated separately in accordance
with the treatment arms. MR4.0 rates were higher in the group
expressing the e14a2 transcript than the e13a2 transcript group in the
three treatment arms, which included imatinib at a dose of 400 mg plus
IFNα, imatinib at a dose of 400 mg plus cytarabine, and imatinib at a
dose of 800 mg (p<0.001, p=0.004, and p=0.028, respectively).[23]
In
the MDACC study that included 481 patients with chronic phase CML
(CML‐CP), the authors assessed the prognostic significance of
transcripts in four groups of patients treated frontline with different
TKI therapies (imatinib at a dose of 400 mg, imatinib at a dose of 800
mg, dasatinib 50 mg twice daily or 100 mg daily, and nilotinib 800
mg/day). Complete cytogenetic response (CCyR) rates were inferior in
the e13a2 group treated with imatinib 400 mg daily (77%) compared with
other TKIs (90%‐95%). Regarding molecular responses, the CI of MMR and
MR4.5 were significantly superior in the e14a2 and co-expression groups
than the e13a2 group in all of the treatment arms (p<0.001 and
p<0.001, respectively). When treatment responses were assessed for
each specific TKI treatment option, patients with the e13a2 transcript
who had received imatinib 400 mg daily showed a significantly lower
rate of CCyR, MMR, and MR4.5 than those reported among both patients
expressing the e13a2 transcript receiving other TKI treatments and
patients harboring different transcript types receiving imatinib 400 mg
daily. After a long-term follow-up of 60 months, higher CCyR, MMR, and
MR4.5 responses persisted in the group of patients with the e14a2
transcript. In contrast, the MR4.5 response sustainability was lower in
patients with the e13a2 transcript than those with the e14a2 transcript
and transcript co-expression (p<0.001).[24]
An
Italian study conducted by the GIMEMA group including a large cohort of
559 patients treated with imatinib frontline observed that MMR rates at
18 months and MR 4.0 rates at 36 months were significantly inferior in
patients expressing the e13a2 transcript (52% vs. 67%, p=0.001, and 20%
vs. 30%, p=0 .013, respectively). The median time to MMR in the e14a2
and e13a2 groups was 12 and 6 months, with 83% and 88% estimated
probability of achieving MMR (p<0.001), respectively. The median
time of attaining MR4.0 was 61 and 41 months, and the estimated rate of
MR4.0 was 52% and 67% in the two classes of patients (p=0.001),
respectively.[25]
Lin at al. retrospectively
analyzed a cohort of 166 patients (36.7% of patients had e13a2
transcripts, 50% had e14a2, and 13.3% co-expressed e13a2 and e14a2 at
baseline) treated for up to 10 years, focusing on the correlation
between BCR‐ABL1 transcript
type and molecular responses to imatinib after a long-term follow-up.
Patients with e14a2 or both e14a2 and e13a2 transcripts had higher MMR
rates than those with e13a2 (81.8% vs 60.7% [p=0.023] for e14a2 vs
e13a2, respectively, and 77.1% vs 60.7% (p=0.043) for both transcripts
vs e13a2, respectively). The median time to achieve MMR, disease
progression rates and the median time to disease progression did not
differ between the three groups.[27]
A Korean
study evaluated outcomes in patients who received imatinib frontline
with EMR failure at three months to individualize potential predictive
factors for an overall MMR. In this specific subset of patients,
multivariate analyses showed that a transcript type of e13a2 compared
with e14a2 and larger spleen size (>9 cm spleen size) represented
independent risk factors for failure of overall MMR. According to these
results, the authors identified a high-risk group of patients with the
previously cited features who would benefit from early decision-making
regarding treatment change.[29] Another study by the
MDACC group assessed responses to imatinib according to the transcript
not only in 251 patients who received imatinib frontline but also in
229 patients treated with imatinib after IFN‐α failure. The CCyR rates
were similar for patients with e14a2 and e13a2 in both the newly
diagnosed (91% and 82%) and post-IFN failure (72% and 78%) groups. The
rates of MMR and complete molecular response (CMR) (defined as
undetectable transcript levels) were significantly higher in patients
who harbored the e14a2 transcript than those with the e13a2 transcript
(59% vs. 77%; p=0.008 and 25% vs. 47%; p=0.002, respectively) in the
treatment‐naive group. Similar results were also found among the group
of patients who had failed IFNα long-term treatment; the rates of MMR
and CMR were superior in patients with the e14a2 transcript than those
with the e13a2 transcript (34% vs. 63%; p=0.001 and 16% vs. 42%;
p=0.001, respectively).[30]
Among studies that
investigated the impact of transcripts on imatinib response, only one
conducted by an Indian group showed better responses in patients with
e13a2 than those with e14a2. In this study, the CCyR rates were
significantly higher in patients with e13a2 transcripts (59% vs. 28%;
p=0.04). However, in the cohort of 70 patients analyzed, some patients
(n=40) had received previous treatment with hydroxyurea or IFNα.
Therefore, the authors analyzed only the cohort of treatment-naive
patients and reported similar CCyR rates among different transcript
groups (p=0.396)[31] (Table 2 and Table 3).
According to these data, patients with e13a2 transcripts treated with
imatinib had a lower and more slow achievement of CCyR, MMR, and DMR
than those with e14a2 transcripts. They probably should be considered a
high-risk group who would most benefit from treatment with 2GTKIs. To
date, the main endpoint of TKIs has become the achievement of s-DMR,
allowing discontinuation of therapy. Therefore patients with e13a2
would probably require a more potent frontline therapy able to induce
more profound and faster molecular responses. Data on responses to
imatinib in patients with co-expression of both transcripts remain
controversial. Still, most of the studies documented that this group of
patients seems to have better responses and prognoses compared to the
e13a2 group.
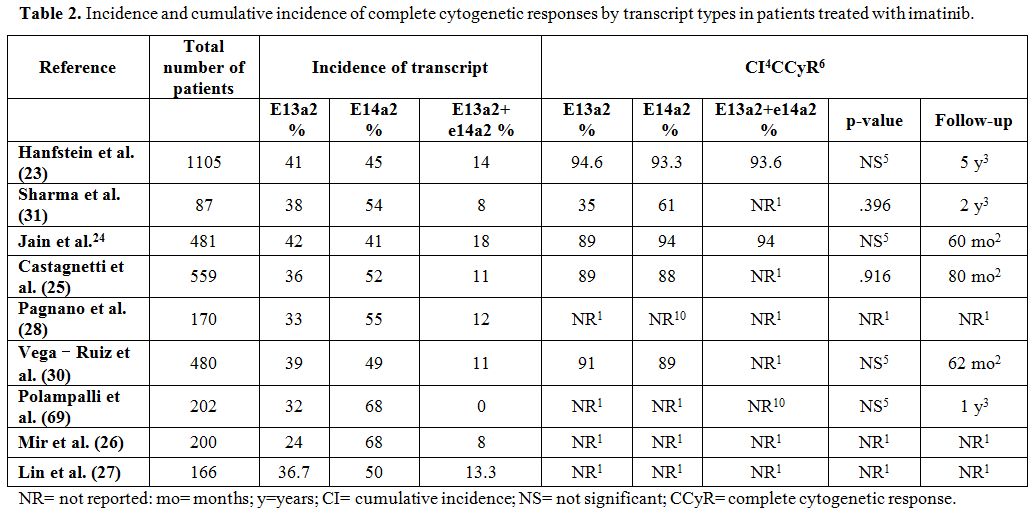 |
Table 2. Incidence and cumulative
incidence of complete cytogenetic responses by transcript types in
patients treated with imatinib. |
 |
Table 3 Cumulative incidence of molecular responses by transcript types in patients treated with imatinib. |
Type of Transcript and Response to 2GTKIs
The
2GTKIs dasatinib and nilotinib have increased the cytogenetic and
molecular response rates in CML patients when used as a frontline
approach or as second-line therapy after imatinib failure for
resistance or intolerance.[32,33,34] To the best of
our knowledge, few previously published studies have systematically and
retrospectively analyzed response rates in patients who received 2GTKIs
frontline according to the type of BCR-ABL1 transcript detected at
diagnosis.
The MDACC study included 105 patients who had received
dasatinib (50 mg twice daily or 100 mg daily) and 108 patients who had
received nilotinib (400 mg twice daily) as frontline treatment.
Patients with the e13a2 transcript in both 2GTKI groups achieved
overall CCyR rates superior to that of imatinib at a dose of 400 mg/day
(95% vs. 77%), but similar to that of imatinib at a dose of 800 mg/day
(95% vs. 90%). Similarly, for MMR and MR4.5, the e13a2 group who
received imatinib 400 mg daily showed a trend of a lower response rate
compared with the groups of patients treated with other TKI approaches.
According to the MMR and MR4.5 response rates, they were substantially
comparable in all TKI modalities for patients expressing e14a2
transcripts except for those who received nilotinib, who had a lower
rate of MR4.5 in both the e13a2 and e14a2 cohorts compared with
patients treated with imatinib 800 mg daily or dasatinib 50 or 100 mg
daily. In fact, the MR4.5 rate in patients with e14a2 transcripts
treated with nilotinib 400 mg twice daily was inferior compared to the
patients with e14a2 who received imatinib 800 mg/day and dasatinib 50
mg twice daily or 100 mg/day (64% in the group of patients treated with
imatinib 400 mg/day, 85% in the group who received imatinib 800 mg/day,
89% in the group who received dasatinib 50 mg twice daily or 100
mg/day, and 68% in the group treated with nilotinib 800 mg/day).
In
contrast, the CCyR and MMR response rates in patients with the e14a2
transcript treated with frontline nilotinib were comparable to those of
other treatment arms. The authors also concluded that patients
receiving 2GTKIs who expressed e14a2 had a trend in favor of achieving
more rapid and deeper cytogenetic and molecular responses than those
with e13a2 transcripts and were able to maintain these responses for a
long time. Furthermore, they also added that expressing the e14a2
transcript (compared with patients harboring e13a2 transcripts but not
the co-expressing patients), receiving frontline imatinib at a dose of
800 mg/day or 2GTKIs, and presenting a spleen size <10 cm at
diagnosis represented prognostic factors for EFS. They also reported
that expressing the e14a2 transcript or co-expressing the e13a2 plus
e14a2 transcripts significantly increased the probabilities of
achieving MMR at six months and 12 months of TKIs treatment. In a
multivariate analysis, positive predictors for TFR were first-line
treatments with imatinib 800 mg daily or dasatinib 50 mg twice daily or
100 mg/day and having the e14a2 transcript or co-expressing the e13a2
plus e14a2 transcripts.[24]
The Italian GIMEMA group assessed whether the BCR-ABL1
transcript type (e14a2 vs. e13a2) affected responses and clinical
outcomes in 345 newly diagnosed adult patients treated frontline with
nilotinib. The response and outcome rates were uniformly lower in the
group of patients with e13a2 transcripts (N=124) than the group with
e14a2 transcripts (N=174), but these differences were not statistically
significant: MMR by 12 months, 66% vs 72%, p=0.244; MR4.0 by 36 months,
56% vs 66%, p=0.067; estimated CI of MMR, 82% vs 88%, p=0.135;
estimated CI of MR4.0, 60% vs 69%, p=0.101; estimated PFS, 88% vs 93%,
p=0.547; and estimated OS, 89% vs 94%, p=0.436. The responses of
patients who co-expressed the e13a2 and the e14a2 transcripts (N=30)
were comparable to those of e14a2 patients.[35]
In
another MDACC study, the authors assessed whether the transcript type
might affect responses to ponatinib. The analysis included 85 patients
(47 with recurrent/refractory CML and 38 newly diagnosed patients)
treated with ponatinib. Among recurrent/refractory patients, responses
to e13a2 and e14a2 and both transcripts were CCyR 50% vs 61% vs 50% and
MMR 29% vs 52% vs 30%, respectively. Among patients in the frontline
setting, the median levels of transcripts at three months were 0.098,
0.091, and 0.042 in patients with e13a2, e14a2a2, and both transcripts,
respectively, and therefore, no differences in terms of responses were
documented.[36]
In patients treated with second-
and third-generation tyrosine kinase inhibitors, the type of BCR/ABL1
transcript did not seem to affect cytogenetic and molecular responses,
and the presence of e13a2 transcript was not considered an unfavorable
factor for response achievement and time to response. However, further
studies in larger patient cohorts are required to clarify these
findings.
Impact of BCR‐ABL1 Transcript Type on the Achievement of Stable Deep Molecular Responses
A
stable deep molecular response (s-DMR) in CML patients is a
prerequisite for possible discontinuation. Patients reaching a
transcript level of ≤0.01% achieved a 4-log reduction (MR4), whereas a BCR-ABL1/ABL
ratio of ≤0.0032% identified a 4.5-log reduction (MR4.5); both
identified a DMR. Several studies identified biologic features
associated with the probability of achieving a DMR as a stable response
(s-DMR for at least two years and treatment duration with TKIs ≥3
years).[24,37,38,39,40]
The
MDACC group reported that patients with e14a2 and co-expressed
transcripts had a significantly higher probability of achieving a
stable MR4.5 than those with e13a2 (8-year probability, 43% vs. 24%;
p=0.0021).[24]
An Italian cooperative group
correlated the presence of e14a2 transcript types at baseline with a
higher frequency of s-DMR (63% vs. 53%; p=0.07) in 320 patients who had
received imatinib, but a group of patients included in the study had
previously been treated with IFN and the analysis considered only MR4
responses and not MR4.5 response rates.[37]
Our
Italian group reported that in univariate analysis (43% vs. 31%,
p=0.02) and multivariate regression analysis (e14a2 vs. e13a2 type, HR
1.6, 95% CI: 1.3-2.9; p=0.03), the e14a2 type of transcript was
associated with higher achievement of a stable MR4.5 compared to the
e13a2 transcript in a series of 208 patients treated with imatinib
frontline.[38] An Australian group showed that in a
series of 298 patients, 48% of patients with e14a2 transcripts were
candidates for a TFR attempt compared with only 32% of e13a2
transcripts after an 8-year follow-up.[39]
In
another recent Italian study comparing patients who had achieved s-DMR
and patients who did not achieve it, the authors did not find any
significant difference according to sex, age, Sokal score distribution,
frontline TKI treatment (imatinib vs. 2GTKIs), and duration of TKI
treatment. Still, the type of BCR-ABL1
transcript was the only baseline characteristic that significantly
predicted the potential achievement of s-DMR. Indeed, the e14a2
transcript was detected at diagnosis in 56/75 (75%) s-DMR-positive
patients and in 29/59 (49%) s-DMR-negative patients (p =0.0023)[40] (Table 4).
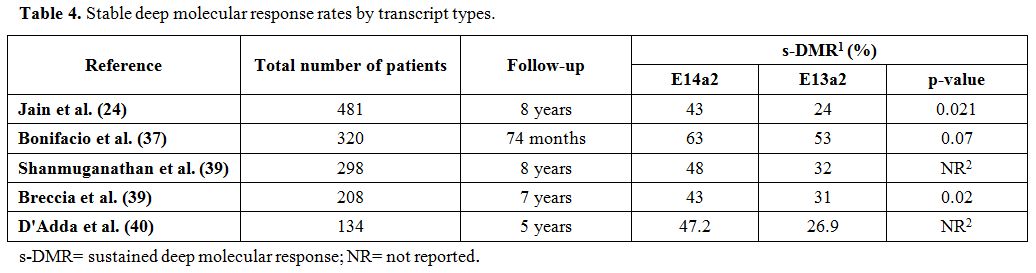 |
Table 4. Stable deep molecular response rates by transcript types. |
The
results of these studies conducted in real-life settings indicated that
the identification of the type of transcript at baseline might help to
identify better those patients who are more likely to benefit from
therapy discontinuation strategies.
Impact of BCR‐ABL1 Transcript Type on TFR
Treatment-free
remission (TFR) is defined as the interval between the date of
discontinuing TKI treatment and that of documented molecular relapse or
if this did not happen, the date of the last follow-up. The TFR is a
new endpoint for CML patients receiving TKIs, with approximately 40%
s-DMR after discontinuing treatment.[41,42] However, precisely predicting who will achieve TFR and the subjects remains a difficult challenge.
The Hammersmith group investigated the correlation between the type of BCR-ABL1
transcript and the probability of TFR in 64 CML patients (37 patients
with the e14a2 transcript and 27 patients with the e13a2 transcript)
who stopped TKI therapy maintaining MR4.0 or MR4.5 for at least 12
months. At the time of stopping TKI, 32 patients were receiving
imatinib and 32 nilotinib or dasatinib. Thirty-seven patients (58%)
remained in molecular remission at a median time of 26 months (range
7-64 months) after discontinuing TKI treatment, and presenting the
e14a2 BCR-ABL1 transcript was
significantly associated with superior probabilities of remaining in
TFR compared with the e13a2 transcript (70% vs. 45%). The 3‐year chance
of TFR was 53% for the entire cohort, but patients with e14a2
transcripts had a higher 3‐year probability of TFR than those with
e13a2 transcripts (66% vs. 38%). The authors also found that the e14a2
transcript type (p=0.016) and age at diagnosis of 40 years or over
(p=0.003) were the only factors significantly associated with TFR.[43]
In an Australian study including 82 patients, the most relevant finding was that patients eligible for TFR expressing e14a2 BCR-ABL1
transcripts were more likely to maintain TFR at 12 months than those
with e13a2 transcripts (65% vs. 34%; p=0.008) and that patients with
either e14a2 or both transcript types were 2.24 times more likely to
remain in TFR at 12 months than those with e13a2 transcripts. The
authors hypothesized that the higher rate of TFR in the e14a2
transcript group might have been associated with a longer time in MR4.5
before discontinuation (4.1 years in the e14a2 cohort vs. 3.01 years in
the e13a2 group).[39]
A recent Italian study showed that the type of BCR-ABL1
transcript had a significant impact not only on the achievement of
s-DMR but also on the maintenance of TFR. In fact, analyzing the DMR
loss rate after 12 months from TKI discontinuation, the authors found
that patients with e14a2 transcripts had a higher probability of
maintaining TFR than those with e13a2 transcripts (79% vs. 40%;
p=0.012)40 (Table 5).
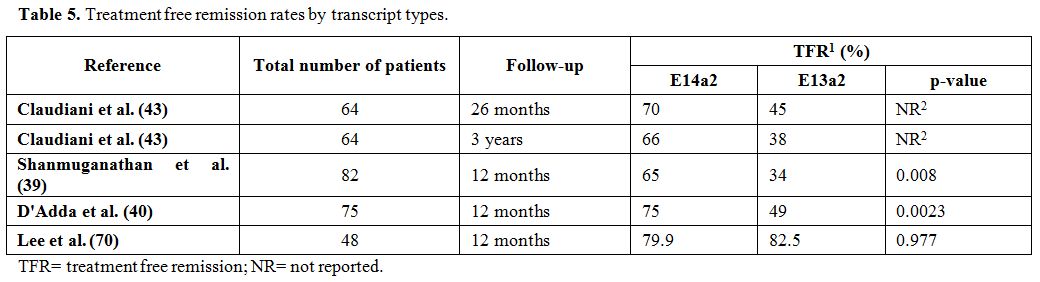 |
Table 5. Treatment free remission rates by transcript types. |
According to these data, having an e13a2 type of BCR-ABL1
transcript is an adverse prognostic factor for achieving s-DMR and
maintaining TFR, while presenting the e14a2 transcript is a favorable
predictive factor for achieving s-DMR, regardless of the TKI type
received and is associated with a consistent rate of TFR maintenance.
.
Type of Transcript and Long-Term Outcome
Several studies[24,25,28,44] analyzed long‐term outcomes and survival data according to different transcript types in CML patients.
The
Italian group observed that the 7‐year OS (90% vs. 83%, p=0.017), PFS
(89% vs. 81%, p=0.005), and failure‐free survival (71% vs. 54%,
p<0.001) rates were significantly higher in patients with e14a2
transcripts than those with e13a2 transcripts and that the transcript
type might be a predictive factor of survival regardless of the daily
imatinib dose.[25] Indeed, in the MDACC studies,
there were no significant differences in 5‐year EFS and OS comparing
patients with the e13a2, e14a2, and co-expressing transcripts. However,
patients with the e13a2 transcript had a worse transformation‐free
survival rate than those with the e14a2 transcript or co-expressed
e13a2 plus e14a2 transcripts (89%, 95%, and 99%, respectively;
p=0.033).[24]
The German group assessed the
prognostic correlation between transcript type and long-term survival
in 1,494 CML patients who received imatinib. The 5-year incidence of
death for CML was 3%, 5%, and 2% (p=0.190) in patients with e14a2
transcripts, e13a2 transcripts, and both transcripts, respectively.
There was also no significant difference in terms of 5‐year OS
comparing patients with the e13a2, e14a2, and co-expressing transcripts
when patients were analyzed according to their ELTS risk scores at
baseline (89%, 93%, and 93%, respectively; p=0.106).[44] Pagnano et al. observed a higher 10-year OS in patients with e13a2 transcripts than those with e14a2 transcripts (p=0.03) (Table 6), but the authors correlated this significance to the younger age of the patients in the e13a2 cohort.[28]
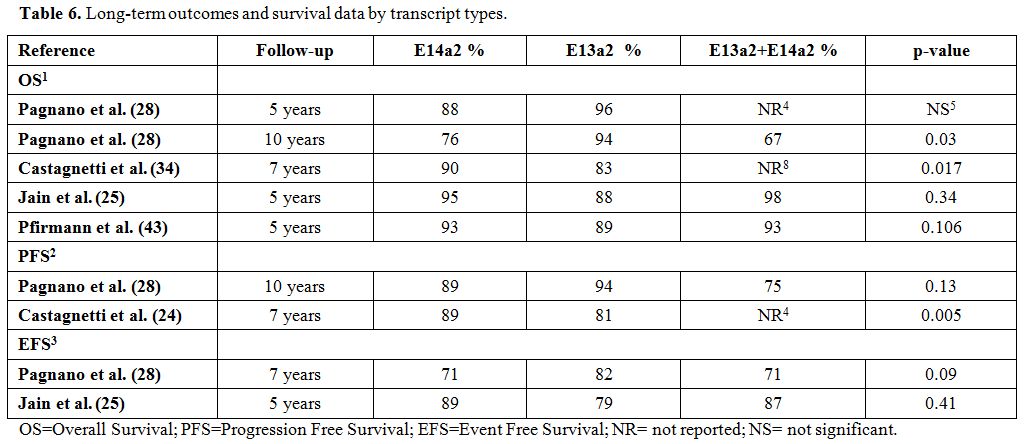 |
Table 6. Long-term outcomes and survival data by transcript types. |
According
to most of the data reported in the literature, the type of transcript
does not seem to affect long-term outcomes of CML patients treated with
TKIs and appears to be a negative prognostic factor when OS, PFS, and
EFS are considered.
Clinical and Prognostic Significance of Atypical BCR-ABL1 Transcript Subtypes in CML
The typical BCR-ABL1
transcript subtypes are e13a2, e14a2, or expression of both
simultaneously, but other less frequently detected transcript subtypes
such as e1a2, e2a2, e6a2, e19a2, e1a3, e13a3, and e14a3 have also been
studied.[45,46] Although there are several published studies on typical BCR-ABL1
fusion transcripts in CML patients from different populations, studies
on the influence of rare transcript subtypes on the disease course and
patient outcomes remain controversial.
The MDACC group assessed
the impact of the e1a2 transcript subtype in a large cohort of 2,322
CML patients treated with TKIs and observed that the incidence of e1a2
transcripts was extremely rare in CML patients (41 patients, 1.8%).
According to the baseline characteristics, CML with e1a2 transcripts
was diagnosed prevalently in older patients (p<0.001) and more
likely presented a blast phase (BP) at diagnosis (p<0.001) compared
to patients with typical transcripts. Furthermore, patients who
expressed e1a2 transcripts showed a higher frequency of additional
chromosomal abnormalities (ACAs) than those with typical transcripts
(46.3% vs. 25.2%, p=0.002). According to treatment responses, patients
with e1a2 transcripts responded more slowly and less likely achieved
CCyR (median time to CCyR 53.1 vs. 18.8 months, p=0.003; overall CCyR
rate 33.3% vs. 66.5%) and MMR (median time to MMR unreached vs. 31.7
months, p=0.001; overall MMR rate 18.5% vs. 63.7%) than those with
typical transcripts. In addition, regarding outcomes, patients with
e1a2 transcripts showed a significantly shorter OS than patients with
typical transcripts, with a median OS of 69.5 vs. 206.8 months
(p<0.001), respectively.[47]
Small series of patients co-expressing e1a2 and e13a2/e14a2 at diagnosis were described regarding responses to TKIs.[48,49]
Our Italian group reported treatment responses and outcomes of 29 CML
patients co-expressing p190 and p210 proteins. In our cohort, after a
median follow‐up of 7 years, median OS was 69 months, and EFS was 69%;
28.5% of patients developed resistance to imatinib, and 14.2%
experienced a BP. Among eight patients who started frontline on
nilotinib, 6 achieved MMR after a median time of 18.8 (range 4-36)
months, and two obtained MR4.5 after three months of therapy. In our
experience, co-expression of e1a2 and a13a2/e14a2 transcripts was
associated with superior rates of resistance and disease progression in
patients who received imatinib, whereas, even in a small cohort of
patients, treatment with 2GTKIs frontline was associated with better
outcomes.[49] Patients with e1a2 transcripts at
diagnosis are rare and associated with a minor issue to therapy with
imatinib. These patients need to be identified as high-risk patients
and receive 2GTKIs as frontline treatment.
The e19a2 rearrangement was initially observed in neutrophilic CML with a benign clinical evolution.[14,50]
Still, it was later reported mainly in patients with typical CML, and
some of these patients exhibited an aggressive clinical course.
According to the published literature, approximately 50 patients with
e19a2 BCR-ABL1 have been reported in CML, and among them, 16 patients received TKIs.[51,52,53,54,55,56]
Of the 16 patients, 13 received imatinib as frontline treatment; among
them, six patients achieved CCyR, and 2 had the first MMR with imatinib
and second MMR with dasatinib. Three out of 13 patients did not respond
to imatinib. One out of 13 patients achieved first MCyR with imatinib
and second with nilotinib, while one patient did not respond to
imatinib but reached MCyR with dasatinib. Therefore, patients with
e19a2 seem to have better responses to 2TKIs.
The e1a3
CML-related atypical translocation is associated with an indolent
clinical course, low leukocyte count, long duration of chronic phase
even without treatment, and a good rate of responses to TKIs.[57,58]
However, Martinez-Serra et al. reported a case of an e1a3-positive
patient who, after an initial response to imatinib, experienced a
lymphoid blast crisis.[59]
In the literature,
fewer than 20 CML cases were reported in which e6a2 fusion was usually
associated with a clinically aggressive disease frequently presenting
in accelerated or blast crisis phases.[60,61,62] Although responses to imatinib have been reported,[63,64] several cases of ABL1 kinase domain mutation-associated imatinib-resistant e6a2 BCR-ABL1 CML have been documented[65,66] with limited information on the efficacy of frontline 2GTKIs in this genotype.
The
prognostic significance of atypical transcripts remains controversial
(except for e1a2) due to different disease genotypes correlated with
each transcript and the small number of patients treated with TKIs.
Conclusions and Future Directions
In
this review, we reported on the influence of the transcript type on
molecular and cytogenetic responses achieved after different TKI
regimens in newly diagnosed CML patients and compared different
transcripts according to survival outcomes and TFR rates.
Several
studies reported that imatinib-treated patients with e14a2 transcript
(and to some extent those with co-expression of e14a2 and e13a2)
obtained more rapid and deeper cytogenetic and molecular responses than
those with only e13a2 transcripts and maintained these responses
longer.[24,25,28] However, in the
majority of studies, e14a2 transcripts did not seem to be associated
with better outcomes in terms of long-term OS, EFS, and PFS[24,28,44] in patients who received imatinib frontline. Therefore, the e13a2 BCR‐ABL1
transcript negatively affects the rate, depth, and speed of responses
to imatinib, and including the transcript type in the calculation of
the baseline risk scores may improve prognostic stratification and
assist with choosing the best treatment policy.
Scant data on the prognostic influence of the BCR-ABL1
transcript type in CML patients treated frontline with second- and
third-generation TKIs are available. In this setting, although a trend
in lower response rates and inferior outcomes in patients with e13a2
transcripts has been reported, the observed differences were
predominantly not significant between e13a2 and e14a2 groups.[35,36]
Further studies of larger patient cohorts are required to clarify
whether 2GTKIs are able to overcome the adverse prognostic impact of
transcript type, potentially improving the probability of achieving
s-DMR and TFR rates and patient outcomes. To date, patients with e13a2
transcripts, if possible (no cardiovascular comorbidities or previous
respiratory diseases), could be considered for treatment with 2GTKIs
or, if patients are not eligible for 2GTKIs due to baseline
comorbidities, the molecular monitoring should be conducted more
strictly and carefully. However, the new version of ELN guidelines[71]
does not recommend any specific treatment choice according to the type
of transcript at baseline. The type of transcript has not yet been
included in the prognostic scores generally used for patients with CML
and age, comorbidities, and EUTOS Long Term Survival (ELTS) risk-score
at diagnosis remain the main factors that guide the therapeutic
strategy.
For CP-CML patients, the TFR is increasingly becoming
a goal of therapy; however, the ability to predict success following
attempted TFR remains limited. The new ELN guidelines[71] require the presence of typical e13a2 or e14a2 BCR–ABL1 transcripts for potentially attempting TFR. Recent studies[38,39,40,43] found that the e14a2 BCR-ABL1
transcript was significantly associated with a higher rate of TFR
regardless of the TKI used. Furthermore, it was also observed that
patients expressing e14a2 transcripts have a considerably higher
incidence of stable MR4.5 response than those with e13a2 transcripts.[25,38,39,40]
Therefore, the type of transcript may also increase the probability of
reaching the endpoint required for treatment discontinuation.
Among
atypical transcripts potentially associated with CML, the e1a2
transcript deserves particular attention. Patients with e1a2
transcripts are diagnosed at an older median age and are more likely to
present in BP initially; those who do not present in BP at baseline
have an increased risk of subsequent progression to BP, lower
cytogenetic and molecular responses to TKI treatments, and dismal OS.[47,48]
This transcript is a high-risk factor for disease progression, and
patients should always be considered for frontline 2GTKI treatment.
References
- Jabbour E, Kantarjian H. Chronic myeloid leukemia:
2018 update on diagnosis, therapy and monitoring. Am J Hematol. 2018;
93(3):442-459. https://doi.org/10.1002/ajh.25011 PMid:29411417
- Rowley
JD. A new consistent chromosomal abnormality in chronic myelogenous
leukaemia identified by quinacrine fluorescence and Giemsa staining.
Nature. 1973; 243(5405):290-293. https://doi.org/10.1038/243290a0 PMid:4126434
- Kurzrock
R, Gutterman JU, Talpaz M. The molecular genetics of Philadelphia
chromosome-positive leukemias. N Engl J Med. 1988; 319(15):990-8. https://doi.org/10.1056/NEJM198810133191506 PMid:3047582
- Kurzrock
R, Kantarjian HM, Druker BJ, Talpaz M. Philadelphia chromosome-positive
leukemias: from basic mechanisms to molecular therapeutics. Ann Intern
Med. 2003; 138(10):819-30. https://doi.org/10.7326/0003-4819-138-10-200305200-00010 PMid:12755554
- Bennour
A, Ouahchi I, Achour B, Zaier M, Youssef YB, Khelif A, Saad A, Sennana
H. Analysis of the clinico-hematological relevance of the breakpoint
location within M-BCR in chronic myeloid leukemia. Med Oncol. 2013;
30(1):348. https://doi.org/10.1007/s12032-012-0348-z PMid:23269583
- Groffen
J, Stephenson JR, Heisterkamp N, de Klein A, Batram CR, Grosveld G..
Philadelphia chromosomal breakpoints are clustered within a limited
region, bcr, on chromosome 22. Cell. 1984; 36:93-9. https://doi.org/10.1016/0092-8674(84)90077-1
- Quintás-Cardama A, Cortes J. Molecular biology of bcr-abl1-positive chronic myeloid leukemia. Blood. 2009; 113:1619-30. https://doi.org/10.1182/blood-2008-03-144790 PMid:18827185 PMCid:PMC3952549
- Hai
A, Kizilbash NA, Zaidi SH, Alruwaili J, Shahzad K. Differences in
structural elements of Bcr-Abl oncoprotein isoforms in Chronic
Myelogenous Leukemia. Bioinformation. 2014; 10(3): 108-114. https://doi.org/10.6026/97320630010108 PMid:24748748 PMCid:PMC3974235
- Verma
D, Kantarjian HM, Jones D, Luthra R, Borthakur G, Verstovsek S, Rios
MB, Cortes J. Chronic myeloid leukemia (CML) with P190BCR‐ABL: analysis
of characteristics, outcomes, and prognostic significance. Blood. 2009;
114:2232‐2235. https://doi.org/10.1182/blood-2009-02-204693 PMid:19531657 PMCid:PMC4828071
- Winter
SS, Greene JM, McConnell TS, Willman CL. Pre-B acute lymphoblastic
leukemia with b3a2 (p210) and e1a2 (p190) BCR-ABL fusion transcripts
relapsing as chronic myelogenous leukemia with a less differentiated
b3a2 (p210) clone. Leukemia. 1999; 13(12): 2007-11. https://doi.org/10.1038/sj.leu.2401598 PMid:10602422
- Jones
D, Luthra R, Cortes J, Thomas D, O'Brien S, Bueso-Ramos C, Hai S,
Ravandi F, de Lima M, Kantarjian H, Jorgensen JL. BCR-ABL fusion
transcript types and levels and their interaction with secondary
genetic changes in determining the phenotype of Philadelphia
chromosome-positive leukemias. Blood. 2008; 112(13):5190-5192. https://doi.org/10.1182/blood-2008-04-148791 PMid:18809762 PMCid:PMC2597614
- Lichty
BD, Keating A, Callum J, Yee K, Croxford R, Corpus G, Nwachukwu B, Kim
P, Guo J, Kamel-Reid S. Expression of p210 and p190 BCR-ABL due to
alternative splicing in chronic myelogenous leukaemia. Br J Haematol.
1998; 103(3):711-5. https://doi.org/10.1046/j.1365-2141.1998.01033.x PMid:9858221
- Gong
Z, Medeiros JL, Cortes JE, et al. Clinical and prognostic significance
of e1a2 BCR-ABL1 transcript subtype in chronic myeloid leukemia. Blood
Cancer J. 2017; 7(7): e583. https://doi.org/10.1038/bcj.2017.62 PMid:28708130 PMCid:PMC5549254
- Verstovsek
S, Lin H, Kantarjian H, Saglio G, De Micheli D, Pane F, Garcia-Manero
G, Intrieri M, Rotoli B, Salvatore F, Guo JQ, Talpaz M, Specchia G,
Pizzolo G, Liberati AM, Cortes J, Quackenbush RC, Arlinghaus RB.
Neutrophilic-chronic myeloid leukemia: low levels of p230 BCR/ABL mRNA
and undetectable BCR/ABL protein may predict an indolent course. Cancer
2002; 94:2416-2425. https://doi.org/10.1002/cncr.10490 PMid:12015767
- Baccarani
M, Castagnetti F, Gugliotta G, Rosti G, Soverini S, Albeer A, Pfirrmann
M, the International BCR-ABLStudy Group. The proportion of different
BCR-ABL1 transcript types in chronic myeloid leukemia. An international
overview. Leukemia. 2019. https://doi.org/10.1038/s41375-018-0341-4 PMid:30675008
- Chasseriau
J, Rivet J, Bilan F, Chomel JC, Guilhot F, Bourmeyster N, Kitzis A.
Characterization of the Different BCR-ABL Transcripts with a Single
Multiplex RT-PCR. J Mol Diagn. 2004; 6(4): 343-347. https://doi.org/10.1016/S1525-1578(10)60530-2
- Burmeister
T, Reinhardt R. A multiplex PCR for improved detection of typical and
atypical BCR-ABL fusion transcripts. Leuk Res. 2008; 32(4):579-85. https://doi.org/10.1016/j.leukres.2007.08.017 PMid:17928051
- Limsuwanachot
N, Siriboonpiputtana T, Karntisawiwat K, Chareonsirisuthigul T,
Chuncharunee S, Rerkamnuaychoke B. Multiplex RT-PCR Assay for Detection
of Common Fusion Transcripts in Acute Lymphoblastic Leukemia and
Chronic Myeloid Leukemia Cases. Asian Pac J Cancer Prev. 2016;
17(2):677-84. https://doi.org/10.7314/APJCP.2016.17.2.677 PMid:26925663
- Mills
KL, Mackenzie ED, Birnie GD. The site of the breakpoint within the bcr
is a prognostic factor in Philadelphia chromosome‐positive chronic
myeloid leukemia. Blood. 1989; 72:1237-1241. https://doi.org/10.1182/blood.V72.4.1237.1237
- Schaefer-Rego
K, Dudek H, Popenoe D, Z Arlin, Mears JG, Bank A, Leibowitz D. CML
patients in blast crisis have breakpoints localized to a specific
region of the BCR. Blood. 1987; 70(2):448-55. https://doi.org/10.1182/blood.V70.2.448.448 PMid:3038213
- Shepherd
P, Suffolk R, Halsey J, Allan N. Analysis of molecular breakpoint and
m‐RNA transcripts in a prospective randomized trial of interferon in
chronic myeloid leukaemia: no correlation with clinical features,
cytogenetic response, duration of chronic phase, or survival. Br J
Haematol. 1995; 89:546-554. https://doi.org/10.1111/j.1365-2141.1995.tb08362.x PMid:7734353
- The
Italian Cooperative Study Group on Chronic Myeloid Leukemia. Chronic
myeloid leukemia, BCR/ABL transcript, response to interferon, and
survival. Leukemia. 1995; 9:1648-1652.
- Hanfstein
B, Lauseker M, Hehlmann R, Saussele S, Erben P, Dietz C, Fabarius A,
Proetel U, Schnittger S, Haferlach C, Krause SW, Schubert J, Einsele H,
Hänel M, Dengler J, Falge C, Kanz L, Neubauer A, Kneba
M, Stegelmann F, Pfreundschuh M, Waller CF, Spiekermann K,
Baerlocher GM, Pfirrmann M, Hasford J , Hofmann WK , Hochhaus A, Müller
MC, SAKK and the German CML Study Group. Distinct characteristics of
e13a2 versus e14a2 BCR‐ABL1 driven chronic myeloid leukemia under
first‐line therapy with imatinib. Haematologica. 2014; 99:1441‐1447. https://doi.org/10.3324/haematol.2013.096537 PMid:24837466 PMCid:PMC4562532
- Jain
P, Kantarjian H, Patel KP, Gonzalez GN, Luthra R, Shamanna RK, Sasaki
K, Jabbour E, Romo CG, Kadia TM, Pemmaraju N, Daver N, Borthakur G,
Estrov Z, Ravandi F, O'Brien S, Cortes J. Impact of BCR‐ABL transcript
type on outcome in patients with chronic‐phase CML treated with
tyrosine kinase inhibitors. Blood. 2016; 127:1269‐1275. https://doi.org/10.1182/blood-2015-10-674242 PMid:26729897 PMCid:PMC4786836
- Castagnetti
F, Gugliotta G, Breccia M, Iurlo A, Levato L, Albano F, Vigneri P,
Abruzzese E, Rossi G, Rupoli S, Cavazzini F, Martino M, Orlandi E,
Pregno P, Annunziata M, Usala U, Tiribelli M, Sica S, Bonifacio M, Fava
C, Gherlinzoni F, Bocchia M, Soverini S, Bochicchio MT, Cavo M ,
Martinelli G, Saglio G, Pane F, Baccarani M, Rosti G, GIMEMA CML
Working Party. The BCR‐ABL1 transcript type influences response and
outcome in Philadelphia chromosome-positive chronic myeloid leukemia
patients treated frontline with imatinib. Am J Hematol. 2017;
92:797‐805. https://doi.org/10.1002/ajh.24774 PMid:28466557
- Mir
R, Ahmad I, Javid J, Zuberi M, Yadav P, Shazia R, Masroor M, Guru S,
Ray PC, Gupta N, Saxena A. Simple multiplex RT‐PCR for identifying
common fusion BCR‐ABL transcript types and evaluation of molecular
response of the a2b2 and a2b3 transcripts to imatinib resistance in
north Indian chronic myeloid leukemia patients. Indian J Cancer. 2015;
52:314‐318. https://doi.org/10.4103/0019-509X.176741 PMid:26905124
- Lin
HX, Sjaarda J, Dyck J, Stringer R, Hillis C, Harvey M, Carter R,
Ainsworth P, Leber B, Pare G, Sadikovic B. Gender and BCR‐ABL
transcript type are correlated with molecular response to imatinib
treatment in patients with chronic myeloid leukemia. Eur J Haematol.
2016; 96:360‐366. https://doi.org/10.1111/ejh.12597 PMid:26059983
- Pagnano
KBB, Miranda EC, Delamain MT, Oliveira Duarte G, de Paula EV,
Lorand-Metze I, de Souza GA.Influence of BCR‐ABL transcript type on
outcome in patients with chronic‐phase chronic myeloid leukemia treated
with imatinib. Clin Lymphoma Myeloma Leuk. 2017; 17:728‐733. https://doi.org/10.1016/j.clml.2017.06.009 PMid:28822797
- Sung-Eun
L, Soo-Young C, Soo-Hyun K, Hye-Young S, Hae-Lyun Y, Mi-Young L,
Hee-Jeong H, Ki-Hoon K, Kyung-Mi K, Eun-Jung J, Dong-Wook K. Baseline
BCR‐ABL1 transcript type of e13a2 and large spleen size are predictors
of poor long‐term outcomes in chronic phase chronic myeloid leukemia
patients who failed to achieve an early molecular response after 3
months of imatinib therapy. Leuk Lymphoma. 2018; 59:105‐113. https://doi.org/10.1080/10428194.2017.1320711 PMid:28540759
- Vega‐Ruiz
A, Kantarjian H, Shan J, Wierda W, Burger J, Verstovsek S,
Garcia-Manero G, Cortes J. Better molecular response to imatinib for
patients (pts) with chronic myeloid leukemia (CML) in chronic phase
(CP) carrying the b3a2 transcript compared to b2a2 [abstract]. Blood.
2007; 110:1939. https://doi.org/10.1182/blood.V110.11.1939.1939
- Sharma
P, Kumar L, Mohanty S, Kochupillai V. Response to imatinib mesylate in
chronic myeloid leukemia patients with variant BCR‐ABL fusion
transcripts. Ann Hematol. 2010; 89:241‐247. https://doi.org/10.1007/s00277-009-0822-7 PMid:19714331
- Jabbour
E, Kantarjian HM, Saglio G, Steegmann JL, Shah NP, Boqué C, Chuah C,
Pavlovsky C, Mayer J, Cortes J, Baccarani M, Kim DK, Bradley-Garelik
MB, Mohamed H, Wildgust M, Hochhaus A.Early response with dasatinib or
imatinib in chronic myeloid leukemia: 3-year follow-up from a
randomized phase 3 trial (DASISION). Blood 2014; 123(4):494-500. https://doi.org/10.1182/blood-2013-06-511592 PMid:24311723 PMCid:PMC4190618
- Jain
P, Kantarjian H, Nazha A, O'Brien S, Jabbour E, Romo CG, Pierce S,
Cardenas-Turanzas M, Verstovsek S, Borthakur G, Ravandi F,
Quintás-Cardama A, Cortes J. Early responses predict better outcomes in
patients with newly diagnosed chronic myeloid leukemia: results with
four tyrosine kinase inhibitor modalities. Blood 2013;
121(24):4867-4874. https://doi.org/10.1182/blood-2013-03-490128 PMid:23620574 PMCid:PMC3743466
- Larson
RA, Hochhaus A, Hughes TP, Clark RE, Etienne G, Kim DW, Flinn JW,
Kurokawa M, Moiraghi B, Yu R, Blakesley RE, Gallagher NJ, Saglio G,
Kantarjian H.. Nilotinib vs imatinib in patients with newly diagnosed
Philadelphia chromosome-positive chronic myeloid leukemia in chronic
phase: ENESTnd 3-year follow-up. Leukemia 2012; 26(10):2197-2203. https://doi.org/10.1038/leu.2012.134 PMid:22699418
- Catsagnetti
F, Gugliotta G, Breccia M, Stagno F, D'Adda M, Levato L, Carella AM,
Martino B, Tiribelli M, Fava C, Binotto G, Avanzini P, Bocchia M,
Bergamaschi M, Russo Rossi A, Cavazzini F, Abruzzese E, Soverini S,
Alimena G, Cavo M, Martinelli G, Pane F, Saglio G, Baccarani M, Rosti
G. Prognostic Value of BCR-ABL1 Transcript Type in Chronic Myeloid
Leukemia Patients Treated Frontline with Nilotinib. Blood 2016;
128:3070. https://doi.org/10.1182/blood.V128.22.3070.3070
- Jain
P, Kantarjian HM, Romo CG, et al. BCR‐ABL fusion transcripts in
patients with chronic myeloid leukemia in chronic phase (CML‐CP):
experience with ponatinib from MD Anderson Cancer Center [abstract].
Blood. 2013; 122:2727. https://doi.org/10.1182/blood.V122.21.2727.2727
- Bonifacio
M, Scaffidi L, Binotto G, De Marchi F, Maino E, Calistri E, Bonalumi
A,Frison L,Marin L, Medeot M, De Matteis G, Fanin R, Semenzato G,
Ambrosetti A, Tiribelli M. Predictive Factors of Stable Deep Molecular
Response in Chronic Myeloid Leukemia Patients Treated with Imatinib
Standard Dose: A Study from the Gruppo Triveneto LMC. Blood 2015;
126:597. https://doi.org/10.1182/blood.V126.23.597.597
- Breccia
M, Molica M, Colafigli G, Massaro F, Quattrocchi L, Latagliata R, M,
Diverio D, Guarini A, Alimena G, Foà R. Prognostic factors associated
with a stable MR4.5 achievement in chronic myeloid leukemia patients
treated with imatinib. Oncotarget. 2017; 9(7):7534-7540. https://doi.org/10.18632/oncotarget.23691 PMid:29484130 PMCid:PMC5800922
- Shanmuganathan
N, Branford S, Yong ASM, Hiwase DK, Yeung DT, Ross DM, Hughes TP. The
e13a2 BCR-ABL1 Transcript Is Associated with Higher Rates of Molecular
Recurrence after Treatment-Free Remission Attempts: Retrospective
Analysis of the Adelaide Cohort. ASH 2018; Oral Poster 1731. https://doi.org/10.1182/blood-2018-99-111083
- D'Adda
M, Farina M, Cerqui E, Ruggeri G, Ferrari S, Chiara Bottelli C, Pagani
C, Passi A, Rossi G. An e13a2 Type of BCR-ABL Transcript Has a
Significant Adverse Impact on the Achievement of a Sustained Deep
Molecular Response and on the Maintenance of a Treatment Free Remission
after Stopping Tyrosine Kinase Inhibitors. Blood 2017; 130:1589.
- Mahon
FX, Rea D, Guilhot J, Guilhot F, Huguet F, Nicolini F, Legros L,
Charbonnier A, Guerci A, Varet B, Etienne G, Reiffers J, Rousselot P,
Intergroupe Français des Leucémies Myéloïdes Chroniques Discontinuation
of imatinib in patients with chronic myeloid leukaemia who have
maintained complete molecular remission for at least 2 years: the
prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol.
2010; 11(11):1029-1035. https://doi.org/10.1016/S1470-2045(10)70233-3
- Rousselot
P, Charbonnier A, Cony-Makhoul P, Agape P, Nicolini F, Varet B,
Gardembas M, Etienne G, Réa D, Roy L, Escoffre-Barbe M, Guerci-Bresler
A, Tulliez M, Prost S, Spentchian M, Cayuela JM, Reiffers J, Chomel JC,
Turhan A, Guilhot J, Guilhot F, Mahon FX. Loss of major molecular
response as a trigger for restarting tyrosine kinase inhibitor therapy
in patients with chronic-phase chronic myelogenous leukemia who have
stopped imatinib after durable undetectable disease. J Clin Oncol.
2014; 32(5):424-430. https://doi.org/10.1200/JCO.2012.48.5797 PMid:24323036
- Claudiani
S, Apperley JF, Gale RP, Clark R, Szydlo R, Deplano S, Palanicawandar
R, Khorashad J, Foroni L, Milojkovic D.E14a2 BCR‐ABL1 transcript is
associated with a higher rate of treatment‐free remission in persons
with chronic myeloid leukemia after stopping tyrosine kinase‐inhibitor
therapy. Haematologica. 2017; 102:e297‐e299. https://doi.org/10.3324/haematol.2017.168740 PMid:28495914 PMCid:PMC5541883
- Pfirrmann
M, Evtimova D, Saussele S, Castagnetti F, Cervantes F, Janssen J,
Hoffmann VS, Gugliotta G, Hehlmann R, Hochhaus A, Hasford J, Baccarani
M. No influence of BCR‐ABL1 transcript types e13a2 and e14a2 on
long‐term survival: results in 1494 patients with chronic myeloid
leukemia treated with imatinib. J Cancer Res Clin Oncol. 2017;
143:843‐850. https://doi.org/10.1007/s00432-016-2321-2 PMid:28083711
- Lucas
CM, Harris RJ, Giannoudis A, Davies A, Knight K, Watmough SJ, Wang L,
Clark RE. Chronic myeloid leukemia patients with the e13a2 BCR‐ABL
fusion transcript have inferior responses to imatinib compared to
patients with the e14a2 transcript. Haematologica. 2009; 94:1362‐1367. https://doi.org/10.3324/haematol.2009.009134 PMid:19713230 PMCid:PMC2754951
- Burmeister
T, Reinhardt R. A multiplex PCR for improved detection of typical and
atypical BCR-ABL fusion transcripts. Leuk Res 2008; 32: 579-585. https://doi.org/10.1016/j.leukres.2007.08.017 PMid:17928051
- Verma
D, Kantarjian H, Jones D, Luthra R, Borthakur G, Verstovsek S, Rios MB,
Cortes J. Chronic myeloid leukemia (CML) with P190BCR-ABL: analysis of
characteristics, outcomes, and prognostic significance. Blood. 2009;
114(11): 2232-2235. https://doi.org/10.1182/blood-2009-02-204693 PMid:19531657 PMCid:PMC4828071
- Gong
Z, Medeiros LJ, Cortes JE, Zheng L, Khoury JD, Wang W, Tang G, Loghavi
G, Luthra R, Yang W, Kantarjian H, Hu S. Clinical and prognostic
significance of e1a2 BCR-ABL1 transcript subtype in chronic myeloid
leukemia. Blood Cancer J. 2017; 7(7):e583. https://doi.org/10.1038/bcj.2017.62 PMid:28708130 PMCid:PMC5549254
- Molica
M, Zacheo I, Diverio D, Alimena G, Breccia M. Long-term outcome of
chronic myeloid leukaemia patients with p210 and p190 co-expression at
baseline. Br J Haematol. 2015; 169(1):148-50. https://doi.org/10.1111/bjh.13184 PMid:25296902
- Pane
F, Frigeri F, Sindona M, Luciano L, Ferrara F, Cimino R, Meloni G,
Saglio G, Salvatore F, Rotoli B. Neutrophilic-chronic Myeloid Leukemia:
A Distinct Disease With a Specific Molecular Marker (BCR/ABL With C3/A2
Junction). Blood. 1996; 88(7):2410-4. https://doi.org/10.1182/blood.V88.7.2410.bloodjournal8872410 PMid:8839830
- Lee
JJ, Kim HJ, Kim YJ, Lee S, Hwang JY, Kim YL, Kim DW. Imatinib induces a
cytogenetic response in blast crisis or interferon failure chronic
myeloid leukemia patients with e19a2 BCR-ABL transcripts. Leukemia
2004; 18,1539-1540. https://doi.org/10.1038/sj.leu.2403454 PMid:15284852
- Mondal
BC, Majumdar S, Dasgupta UB, Chaudhuri U, Chakrabarti P, Bhattacharyya
S. E19a2 BCR-ABL fusion transcript in typical chronic myeloid
leukaemia: a report of two cases. J Clin Pathol 2006; 59:1102-1103. https://doi.org/10.1136/jcp.2005.029595 PMid:17021137 PMCid:PMC1861751
- Oshikawa
G, Kurosu T, Arai A, Murakami N, Miura O. Clonal evolution with double
Ph followed by tetraploidy in imatinib-treated chronic myeloid leukemia
with e19a2 transcript in transformation. Cancer Genet Cytogenet 2010;
199:56- 61. https://doi.org/10.1016/j.cancergencyto.2010.01.018 PMid:20417871
- Martin
SE, Sausen M, Joseph A, Kingham B. Chronic myeloid leukemia with e19a2
atypical transcript: early imatinib resistance and complete response to
dasatinib. Cancer Genet Cytogenet 2010; 201:133-134. https://doi.org/10.1016/j.cancergencyto.2010.05.012 PMid:20682399
- Bennour
A, Beaufils N, Sennana S, Saad A, Gabert J. E355G mutation appearing in
a patient with e19a2 chronic myeloid leukaemia resistant to imatinib. J
Clin Pathol 2010; 737- 740. https://doi.org/10.1136/jcp.2010.078311 PMid:20702476
- Langabeer
SE, McCarron SL, Carroll P, Kelly J, O'Dwyer MJ, Conneally E. Molecular
response to first line nilotinib in a patient with e19a2 BCR-ABL1
chronic myeloid leukemia. Leuk Res 2012; 35:e169- e170. https://doi.org/10.1016/j.leukres.2011.05.010 PMid:21641037
- Al-Ali
HK, Leiblein S, Kovacs I, Hennig E, Niederwieser D, Deininger MWN. CML
with an e1a3 BCR-ABL fusion: rare, benign, and a potential diagnostic
pitfall. Blood. 2002; 100(3):1092-1093. https://doi.org/10.1182/blood-2002-03-0930 PMid:12130477
- Roman
J, Jimenez A, Barrios M, J Castillejo A, Maldonado J, Torres A. E1A3 as
a unique, naturally occurring BCR-ABL transcript in an indolent case of
chronic myeloid leukaemia. Br J Haematol. 2001; 114(3):635-637. https://doi.org/10.1046/j.1365-2141.2001.02971.x PMid:11552990
- Martinez-Serra
J, del Campo R, Gutierrez A, Antich JL,Ginard M, Durán MA, Bento L, Ros
T, Amat JC, Vidal C, Iglesias JF, Orlinska I, Besalduch J.Chronic
myeloid leukemia with an e1a3 BCR-ABL fusion protein: transformation to
lymphoid blast crisis. Biomark Res. 2014; 2: 14. https://doi.org/10.1186/2050-7771-2-14 PMid:25197554 PMCid:PMC4155769
- Schultheis
B, Wang L, Clark RE, Melo J V. BCR-ABL with an e6a2 fusion in a CML
patient diagnosed in blast crisis. Leukemia. 2003; 17(10):2054-2055. https://doi.org/10.1038/sj.leu.2403079 PMid:14513059
- Colla
S, Sammarelli G, Voltolini S, Crugnola M, Sebastio P, Giuliani N. E6a2
BCR-ABL transcript in chronic myeloid leukemia: is it associated with
aggressive disease? Haematologica. 2004; 89(5):611-613.
- Beel
KA, Lemmens J, Vranckx H, Maertens J, Vandenberghe P. CML with e6a2
BCR-ABL1 transcript: An aggressive entity? Annals of Hematology. 2011;
90(10):1241-1243. https://doi.org/10.1007/s00277-011-1169-4 PMid:21302112
- Popovici
C, Cailleres S, David M, Lafage-Pochitaloff M, Sainty D, Mozziconacci
MJ. E6a2 BCR-ABL fusion with BCR exon 5-deleted transcript in a
Philadelphia positive CML responsive to imatinib. Leuk Lymph 2005;
46(9):1375-1377. https://doi.org/10.1080/10428190500138138 PMid:16109618
- Breccia
M, Cannella L, Diverio D, Streponi P, Nanni M, Stefanizzi C, Natalino
F, Mecarocci S, Alimena G. Isolated thrombocytosis as first sign of
chronic myeloid leukemia with e6a2 BCR/ABL fusion transcript, JAK2
negativity and complete response to imatinib. Leuk Res. 2008;
32(1):177-180. https://doi.org/10.1016/j.leukres.2007.05.022 PMid:17688946
- Schnittger
S, Bacher U, Kern W, Haferlach T, Hertenstein BM Haferlach C. A new
case with rare e6a2 BCR-ABL fusion transcript developing two new
resistance mutations during imatinib mesylate, which were replaced by
T315I after subsequent dasatinib treatment. Leukemia. 2008;
22(4):856-858. https://doi.org/10.1038/sj.leu.2404949 PMid:17851552
- Vefring
HK., Gruber FXE, Wee L, Kelleher E, O'Shea D, Conneally E, Langabeer
SE. Chronic myelogenous leukemia with the e6a2 bcr-abl and lacking
imatinib response: Presentation of two cases. Acta Haematologica. 2009;
122(1):11-16. https://doi.org/10.1159/000230037 PMid:19641300
- Mondal
BC, Bandyopadhyay A, Majumdar S, Mukhopadhyay A, Chandra S, Chaudhuri
U, Chakrabarti P, Bhattacharyya S, Dasgupta UB. Molecular profiling of
chronic myeloid leukemia in eastern India. Am J Hematol. 2006;
81(11):845-849. https://doi.org/10.1002/ajh.20682 PMid:16888785
- Lee
M, Kantarjian H, Talpaz M. Association of the responsiveness to
interferon therapy with the bcr/abl splicing pattern in Philadelphia
chromosome positive chronic myelogenous leukemia. Blood. 1992;
80(suppl.I):210.
- Polampalli S,
Choughule A, Negi N, Shinde S, Baisane C, Amre P, Subramanian PG,
Gujral S, Prabhash K, Parikh P. Analysis and comparison of
clinicohematological parameters and molecular and cytogenetic response
of two Bcr/Abl fusion transcripts. Genet Mol Res. 2008; 7:1138‐1149. https://doi.org/10.4238/vol7-4gmr485 PMid:19048492
- Lee
SE, Choi SY, Bang JH, Kim SH, Jang EJ, Byeun JY, Park JE, Jeon HR, Oh
YJ, Kim HJ, Kim YK, Park JS, Jeong SH, Kim SH, Zang DY, Oh S, Koo DH,
Kim H, Do YR, Kwak JY, Kim JA, Kim DY, Mun YC, Mauro MJ, Kim DW.
Predictive factors for successful imatinib cessation in chronic myeloid
leukemia patients treated with imatinib. Am J Hematol. 2013;
88:449‐454. https://doi.org/10.1002/ajh.23427 PMid:23440689
- Hochhaus
A, Baccarani M, Silver RT, Schiffer C, Apperley JF, Cervantes F, Clark
RE, Cortes JE, Deininger MW, Guilhot F, Hjorth-Hansen H, Hughes TP,
Janssen JJWM, Kantarjian HM, Kim DW, Larson RA, Lipton JH, Mahon FX,
Mayer J, Nicolini F, Niederwieser D, Pane F, Radich JP, Rea D, Richter
J, Rosti G, Rousselot P, Saglio G, Saußele S, Soverini S, Steegmann JL,
Turkina A, Zaritskey A, Hehlmann R. European LeukemiaNet 2020
recommendations for treating chronic myeloid leukemia. Leukemia. 2020;
34(4):966-984. https://doi.org/10.1038/s41375-020-0776-2 PMid:32127639 PMCid:PMC7214240
[TOP]





