MeiWan Cao1, BaoLing Peng2, WanFu Xu1, PeiYu Chen1, HuiWen Li1, Yang Cheng1, Huan Chen1, LiPing Ye1, Jing Xie1, HongLi Wang1, Lu Ren1, LiYa Xiong1, JingNan Zhu3, XiangYe Xu3, LanLan Geng1 and SiTang Gong1.
1 Department
of Gastroenterology, Guangzhou Women and Children’s Medical Center,
Guangzhou Medical University, Guangzhou City, Guangdong Province,
510623, China.
2 Center for child health and mental health, Shenzhen Childen’s Hospital, Shenzhen City, Guangdong Province, China.
3
Department of Hematology, Guangzhou Women and Children’s Medical
Center, Guangzhou Medical University, Guangzhou City, Guangdong
Province, 510623, China.
4 Guangzhou Institute of
Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou
Medical University, Guangzhou City, Guangdong Province, 510623, China.
Correspondence to:
MeiWan Cao, Department of Gastroenterology, Guangzhou Institute of
Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou
Medical University, Guangzhou City, Guangdong Province, 510623, China.
E-mails:
caomeiwanfirst@hotmail.com
Published: July 1, 2023
Received: December 19, 2022
Accepted: June 17, 2023
Mediterr J Hematol Infect Dis 2023, 15(1): e2023040 DOI
10.4084/MJHID.2023.040
This is an Open Access article distributed
under the terms of the Creative Commons Attribution License
(https://creativecommons.org/licenses/by-nc/4.0),
which permits unrestricted use, distribution, and reproduction in any
medium, provided the original work is properly cited.
|
|
Abstract
Myelodysplastic
syndromes (MDS) are a group of heterogeneous myeloid clonal diseases
that are characterized by ineffective bone marrow hematopoiesis. Since
studies have confirmed the significance of miRNAs in ineffective
hematopoiesis in MDS, the current report elucidated the mechanism
mediated by miR-155-5p. The bone marrow of MDS patients was collected
to detect miR-155-5p and to analyze the correlation between miR-155-5p
and clinicopathological variables. Isolated bone marrow CD34+
cells were transfected with lentiviral plasmids that interfere with
miR-155-5p, followed by apoptosis analysis. Finally,
miR-155-5p-targeted regulation of RAC1 expression was identified, as
well as the interaction between RAC1 and CREB, the co-localization of
RAC1 and CREB, and the binding of CREB to miR-15b. As measured,
miR-155-5p was upregulated in the bone marrow of MDS patients. Further
cell experiments validated that miR-155-5p promoted CD34+
cell apoptosis. miR-155-5p could reduce the transcriptional activity of
miR-15b by inhibiting RAC1, dissociating the interaction between RAC1
and CREB, and inhibiting the activation of CREB. Upregulating RAC1,
CREB, or miR-15b could reduce miR-155-5p-mediated apoptosis promotion
on CD34+
cells. Additionally, miR-155-5p could force PD-L1 expression, and this
effect was impaired by elevating RAC1, CREB, or miR-15b. In conclusion,
miR-155-5p mediates PD-L1-mediated apoptosis of CD34+ cells in MDS by RAC1/CREB/miR-15b axis, thereby inhibiting bone marrow hematopoiesis.
|
Introduction
Myelodysplastic
syndromes (MDS) are a myeloid neoplasm characterized by abnormal
differentiation and development of myeloid cells, manifested as
ineffective hematopoiesis, refractory cytopenia, hematopoietic failure,
and a high risk of transformation to acute myeloid leukemia.[1] For
low-risk MDS, anemia, thrombocytopenia, multiple and/or refractory
cytopenias are preferred, while for high-risk diseases, allogeneic HCT
and hypomethylating agents are clinically recommended.[2] Prediction of
prognosis is complicated by the fact that MDS is primarily a disease of
the elderly, with frequent comorbidities and/or a patient population
with environmental or therapeutic exposure to mutagens.[3] Genomics in
MDS is needed today to improve diagnosis, assign classification and
define prognosis, allow patient-tailored treatment, and even provide
strategies to detect minimal residual disease.[4]
Theoretically,
there are complex interactions between genetic and epigenetic factors,
including miRNAs, ranging from clonal hematopoiesis of uncertain
potential to hematological malignancies. It is underlined that miRNAs
with differential expression may represent pathogenic implications of
MDS in part by regulating hematopoiesis, and miR-155-5p is an
upregulated miRNA in MDS.[5] More importantly, miR-155 transcript
maintains a high level in CD34+ MDS
cells[6] and miR-155 can decrease RAC1 protein to regulate MDS
neutrophils migration.[7] RAC1, a member of the Rho GTPases family,
functions in various hematopoietic cell lineages[8] and RAC1 activation
promotes human hematopoietic stem and progenitor cell maintenance.[9]
Here, the work makes a thorough inquiry on the interlink between
miR-155-5p and RAC1 in CD34+ MDS cell apoptosis.
RAC1
can regulate and activate CREB, a key transcription factor regulating
cell cycle, apoptosis, metabolism, DNA repair, etc.[10] Activated CREB
can bind to the corresponding sites of other genes and regulate
transcriptional expression of target genes,[11,12] including
miR-15b.[13] A report discusses the combined loss-of-function of
miR-15/16 drives the pathogenesis of acute myeloid leukemia.[14]
However, whether miR-15b modulates CD34+ MDS cell apoptosis is still an unsolved question.
Taking into consideration all factors, the present study mainly discussed and explained the mechanism of miR-155-5p in CD34+
MDS cells through targeting RAC1 to manipulate CREB/miR-15b
interaction, determining to carry forward molecule-based therapy for
MDS.
Materials and Methods
Clinical sample collection.
Forty cases of bone marrow specimens were obtained from healthy donors
undergoing orthopedic surgery and MDS patients in Guangzhou Women and
Children’s Medical Center, Guangzhou Medical University, respectively.
The current study complied with the ethical standards of the ethics
committee of Guangzhou Women and Children’s Medical Center, Guangzhou
Medical University. In addition, all patients and healthy donors
obtained written informed consent. Patients are classified according to
the World Health Organization Classification 2022. Prognostic impact
was assessed using the Revised International Prognostic Scoring System
(IPSS-R) and Molecular International Prognostic Scoring System
(IPSS-M).[15,16] Table 1 shows patients’ clinical characteristics. Cytogenetic data of MDS are presented in Supplementary Table 1.
 |
- Table
1. Clinical features of patients with MDS.
|
CD34+ cell isolation. Bone marrow specimens were collected in 2 mM EDTA, from which monocytes were isolated using Ficoll-Paque and CD34+ cells were positively selected using the CD34 Bead Kit (Miltenyi Biotec, USA). Before RNA preparation, CD34+ cells were tested by flow cytometry (CD34+ cells make up more than 90% of isolated cells). Purified CD34+ cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum at 37°C and 5% CO2-saturated humidity.
Cell transfection. miR-155-5p overexpression or knockout stable cell system was constructed in CD34+
cells: Lentiviral plasmids, including Lv-miR-155-5p mimic,
Lv-miR-155-5p inhibitor, Lv-RAC1, Lv-CREB, Lv-miR-15b mimic, and the
corresponding empty controls were produced. Then, 293T cells were
conditioned to lentiviral plasmid transfection, and CD34+ cells were
infected with the virus fluid and added with polybrene. The stably
expressed cell clones were screened with puromycin.
MTT. CD34+ cells at 80% confluence were trypsinized, prepared into a single-cell suspension, and seeded into 96-well plates (3 × 103
cells/well). After 72 h, a medium containing 10% MTT solution (5 g/L,
GD Y1317; Guduo Biotechnology Co, Ltd, Shanghai, China) was added for 4
h, and DMSO (D5879 100ML; Sigma Aldrich, USA) was supplemented at 100
μL/well. Optical density values were measured using a microplate reader
(Nanjing Iron & Steel Co, Ltd, Nanjing, Jiangsu, China) at 490 nm.
Flow cytometry. CD34+
cells conditioned to 48-h culture were detached with 0.25% trypsin,
centrifuged (4°C, 1000 rpm, 5 min), and fixed with pre-chilled 70%
ethanol overnight at 4°C. Next, cells were incubated with 10 μL of
RNase enzyme at 37°C for 5 min, stained with 1% propidium iodide
(40710ES03; Qcbio, Shanghai, China) for 30 min, and placed in a
FACSCalibur (BD Bioscience, USA) to test cell cycle by measuring red
fluorescence at 488 nm.
CD34+
cells after 48-h culture were detached with EDTA-free trypsin and
centrifuged (4°C, 1000 rpm, 5 min). Apoptosis was determined by Annexin
V FITC/PI Apoptosis Detection Kit (CA1020; Solarbio Beijing, China). In
detail, cells were washed with a binding buffer, combined with a 1:40
mixture (Annexin V FITC + binding buffer) for 30 min, and reacted with
another mixture of PI and binding buffer for 15 min. Finally, apoptosis
rates were analyzed by a BD FACS Aria (BD Biosciences).
Hoechst staining. CD34+
cells were seeded into 12-well plates for 48 h and combined with
Hoechst 33342 (Sigma-Aldrich) for 1 h. Darkly stained condensed nuclei
indicate apoptosis.[17]
RT-qPCR. Total RNA from CD34+
cells was extracted using the Trizol kit (15596 026; Invitrogen),
reverse transcribed into cDNA using the reverse transcription kit
(K1621; Fermentas, USA), and analyzed by PCR on an ABI 7500 system.
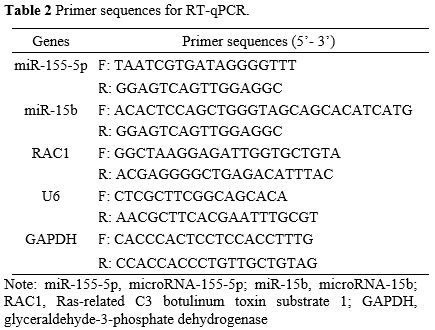
Primer sequences are listed in Table 2 (GeneChem, Shanghai, China). Gene expression was standardized by U6 or GAPDH, and determined by the 2-ΔΔCt method.
 |
- Table
2. Primer sequences for RT-qPCR.
|
Immunoblotting. Protein lysis (R0010; Solarbio) was added to CD34+
cells to isolate total protein, which was then separated using 12%
SDS-polyacrylamide gel electrophoresis (P1200; Solarbio) and loaded
onto a polyethylene difluoride membrane (HVLP04700; Millipore). Next,
nonfat milk-blocked membranes was added with primary antibodies RAC1
(1:1000; 05-389; MilliporeSigma), CREB (1:1000; 9198; Cell Signaling
Technology), p-CREB (1:1000; 9197; Cell Signaling Technology), PD-L1
(1:1000; ab205921) Abcam), Bcl-2 (1:1000; ab32124; Abcam), Bax (1:1000;
ab32503; Abcam) and GAPDH (1:1000; ab8245; Abcam) overnight at 4°C, and
supplemented with horseradish peroxidase-conjugated secondary antibody
(1:2000, ab6721; Abcam) for 2 h. Following diaminobenzidine
development, imaging was done under Gel Doc Xr (Bio-Rad).
Luciferase reporter gene assay.
Constructs with wild-type (WT) or mutant (MUT) miR-155-5p binding sites
were established and designated RAC1-WT or RAC1-MUT. CD34+
cells were seeded into 96-well plates for 24 h and co-transfected with
the constructs, along with miR-155-5p mimic or mimic NC using
Lipofectamine 3000. After 48 h, CD34+ cells were obtained and analyzed using a luciferase reporter gene assay system.
Co-immunoprecipitation (co-IP). CD34+
cells were lysed in HEPES lysis buffer containing 20 mM HEPES (pH 7.2),
50 mM NaCl, 0.5% Triton X-100, 1 mM NaF, 1 mM dithiothreitol and a
cocktail of protease and phosphatase inhibitors. Cell lysates were
incubated with anti-RAC1 or normal rabbit IgG (1:800, N101,
Sigma-Aldrich) for 3 h and added with protein A/G-agarose beads
(SC-2003; Santa Cruz) overnight at 4°C. After washing with HEPES buffer
3 times, immunoprecipitates were collected by 12% SDS-polyacrylamide
gel electrophoresis and tested by immunoblotting.[18]
Immunofluorescence.
Cells were fixed in 4% paraformaldehyde for 20 min and incubated in 50
mM glycine for 20 min. Cells permeabilized in 0.1% Triton X-100 for 30
min were blocked with 10% goat serum (Wako Pure Chemical Industries,
Ltd.) for 60 min, and sequentially incubated with anti-RAC1 and
anti-CREB, followed by co-incubation with Alexa 488-labeled anti-rabbit
IgG antibody (1:1,000, A11031, Thermo Fisher Scientific). After DAPI
staining, images were viewed using a Digital Eclipse C1 TE2000-E
confocal microscope (Nikon).
Chromatin Immunoprecipitation. Following the instructions of ChromaFlash High Sensitivity ChIP Kit (Epigentek), CD34+
cells were transfected with HA-CREB for 24 h, cross-linked with 1%
formaldehyde for 5 min, and added with glycine to 125 mM concentration.
CD34+
cells were sonicated to 100-700 bp, and interacted with HA rabbit
antibody (Abcam), CREB rabbit antibody (Abcam), or rabbit IgG at 4°C.
The extracted and purified ChIP DNA was PCR-amplified with primer
pairs. miR-15b promoter, F: AACCCTTTTCTCACCAGGTCCA; miR-15b promoter,
R: TTCAGAAGAGGGGCGGGCTC. PCR products were separated by electrophoresis
and visualized by UV.
Statistical analysis.
All data were processed by SPSS 21.0, and measurement data were
expressed in the form of mean ± standard deviation. Measurement data
following normal distribution were analyzed by t test, one-way ANOVA
and Tukey's post-hoc test. Survival analysis was implemented with the
Kaplan-Meier test. P < 0.05 represents statistical significance.
Results
miR-155-5p levels in bone marrow CD34+ cells of MDS patients. RT-qPCR found that miR-155-5p was upregulated in the bone marrow CD34+ cells of MDS patients (Figure 1A).
MDS patients were divided into miR-155-5p high expression group and low
expression group according to the median expression of miR-155-5p.
Kaplan-Meier test reported poor survival of MDS patients in the
miR-155-5p high expression group (Figure 1B). Analysis of clinicopathological variables in Table 3
validated that miR-155-5p expression was associated with BM blast
percentage, IPSS-R, IPSS-M, and mutated genes per patient. In the bone
marrow of MDS patients, it tested reduced CD34+
cell proliferation, cell arrest in the G1 phase, increased apoptosis
rates, decreased Bcl-2, and elevated Bax protein expression (Figure 1C-G).
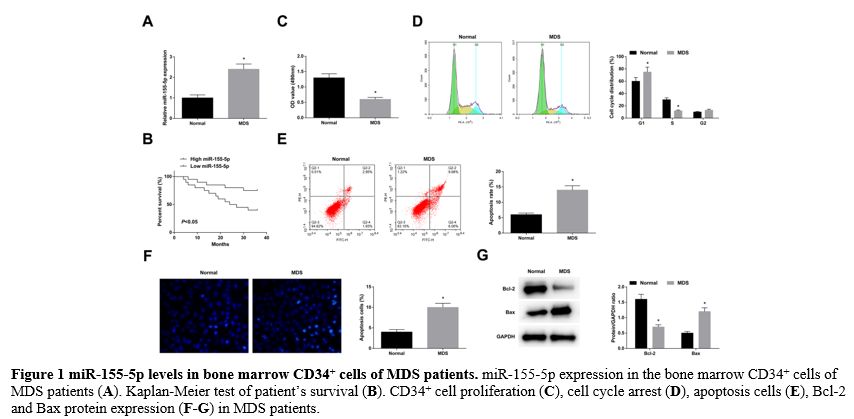 |
Figure 1 miR-155-5p levels in bone marrow CD34+ cells of MDS patients. miR-155-5p expression in the bone marrow CD34+ cells of MDS patients (A). Kaplan-Meier test of patient’s survival (B). CD34+ cell proliferation (C), cell cycle arrest (D), apoptosis cells (E), Bcl-2 and Bax protein expression (F-G) in MDS patients. |
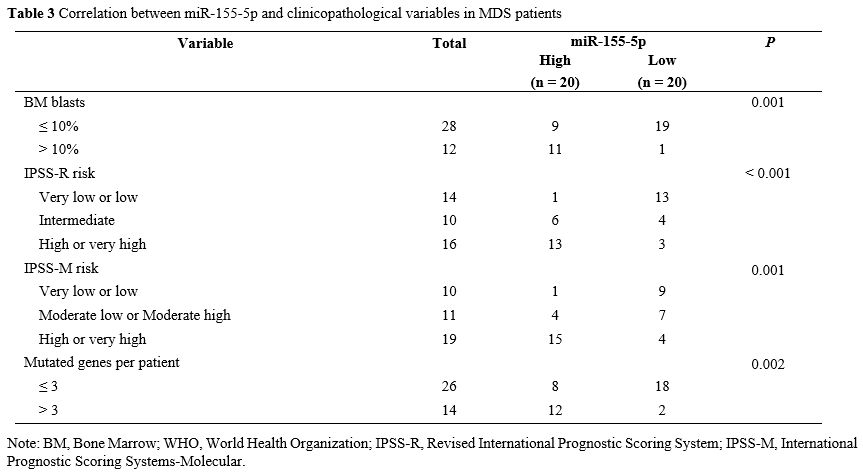 |
Table 3. Correlation between miR-155-5p and clinicopathological variables in MDS patients.
|
miR-155-5p promotes apoptosis of bone marrow CD34+ cells. Lv-miR-155-5p mimic, Lv-miR-155-5p inhibitor, or negative control was transfected into CD34+ cells. RT-qPCR verified that the lentiviral vector was successfully transfected (Figure 2A). MTT proliferation analysis found that after upregulating and downregulating miR-155-5p, CD34+ cell proliferation was obstructed and forced, respectively (Figure 2B). Flow cytometry detection discovered an increased number of CD34+
cells arrested in the G1 phase and promoted apoptosis rates when
miR-155-5p was overexpressed; upon miR-155-5p knockdown, the number of
CD34+ cells in the G1 phase arrest was decreased, along with low apoptosis rates (Figure 2C, D). Hoechst staining manifested that elevating miR-155-5p levels was supportive for CD34+ cell apoptosis, while reducing miR-155-5p levels was suppressive for cellular apoptosis (Figure 2E). Moreover, immunoblotting for apoptosis-related proteins further measured that Bcl-2 decreased and Bax increased in CD34+ cells overexpressing miR-155-5p; the expression trend of the two proteins in CD34+ cells lowly-expressing miR-155-5p were contrary to that in CD34+ cells overexpressing miR-155-5p (Figure 2F). After treatment of CD34+ cells with the Bcl-2 inhibitor venetoclax, cell proliferation decreased and apoptosis increased (supplementary Figure 1A-E), similar to the results after upregulation of miR-155-5p.
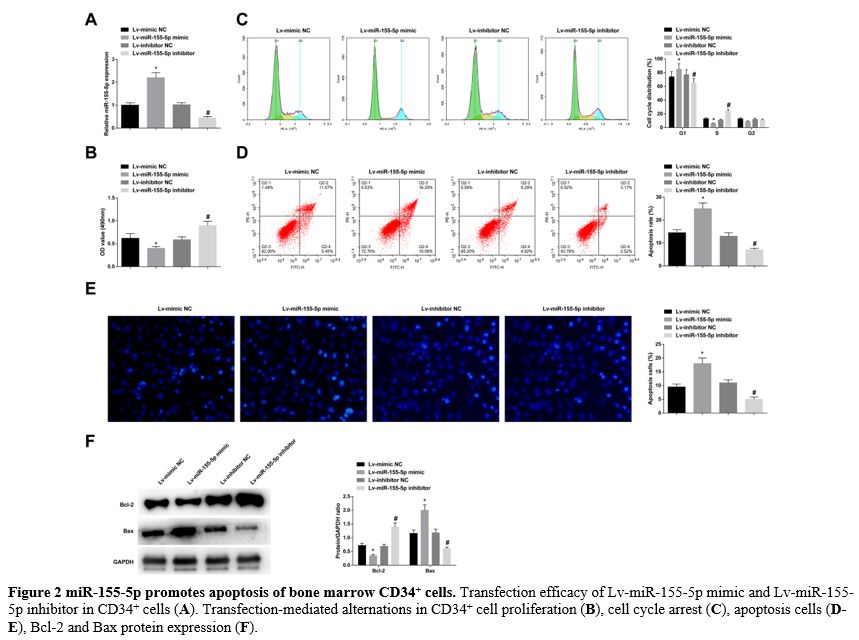 |
- Figure 2 miR-155-5p promotes apoptosis of bone marrow CD34+ cells. Transfection efficacy of Lv-miR-155-5p mimic and Lv-miR-155-5p inhibitor in CD34+ cells (A). Transfection-mediated alternations in CD34+ cell proliferation (B), cell cycle arrest (C), apoptosis cells (D-E), Bcl-2 and Bax protein expression (F).
|
miR-155-5p targets RAC1. Searched by the online software starBase 3.0, it was found a binding site between miR-155-5p and RAC1 (Figure 3A). Luciferase reporter assay supported that miR-155-5p mimic reduced the luciferase activity of the RAC1-WT (Figure 3B). RAC1 was downregulated in bone marrow CD34+ cells of MDS patients (Figure 3C). Moreover, RAC1 expression was enhanced in CD34+ cells transfected with Lv-miR-155-5p mimic, or decreased in those transfected with Lv-miR-155-5p inhibitor (Figure 3D, E).
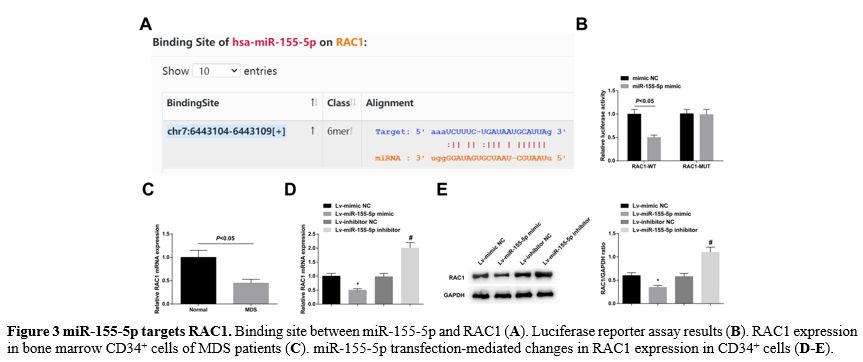 |
- Figure 3 miR-155-5p targets RAC1. Binding site between miR-155-5p and RAC1 (A). Luciferase reporter assay results (B). RAC1 expression in bone marrow CD34+ cells of MDS patients (C). miR-155-5p transfection-mediated changes in RAC1 expression in CD34+ cells (D-E).
|
miR-155-5p modulates CD34+ cell apoptosis by interacting with RAC1. Lv-miR-155-5p mimic + RAC1 were co-transfected into CD34+ cells, and the transfection success was verified (Figure 4A).
In response to RAC1 overexpression, the impacts of miR-155-5p
overexpression came to a reversal, resulting in enhanced proliferation (Figure 4B), cell cycle arrest, reduced apoptosis rates (Figure 4C-E), elevated Bcl-2, and reduced Bax levels (Figure 4F).
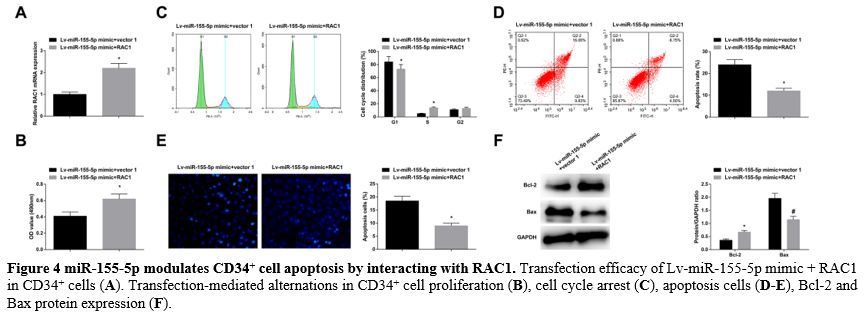 |
- Figure 4 miR-155-5p modulates CD34+ cell apoptosis by interacting with RAC1. Transfection efficacy of Lv-miR-155-5p mimic + RAC1 in CD34+ cells (A). Transfection-mediated alternations in CD34+ cell proliferation (B), cell cycle arrest (C), apoptosis cells (D-E), Bcl-2 and Bax protein expression (F).
|
miR-155-5p reduces the transcriptional activity of miR-15b by RAC1/CREB axis. Immunoprecipitation data validated the interaction of RAC1 with CREB, demonstrating RAC1 binding to CREB (Figure 5A). Then, immunofluorescence staining revealed that RAC1 and CREB co-localized in the nucleus (Figure 5B). Furthermore, chromatin immunoprecipitation found that CREB could bind to the miR-15b promoter (Figure 5C).
It was further measured that elevating miR-155-5p reduced CREB and
p-CREB levels, while downregulating miR-155-5p exerted oppositely (Figure 5D).
In addition, overexpression of miR-155-5p decreased the transcriptional
activity of the miR-15b promoter and miR-15b expression, while
underexpression of miR-155-5p had the contrary effect (Figure 5E, F).
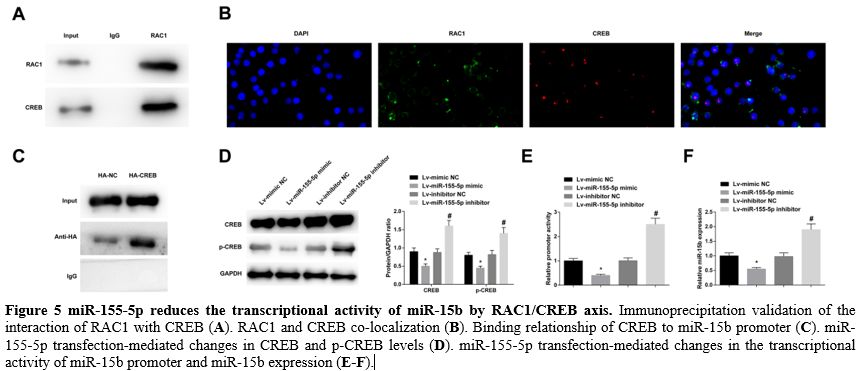 |
- Figure 5 miR-155-5p reduces the transcriptional activity of miR-15b by RAC1/CREB axis. Immunoprecipitation validation of the interaction of RAC1 with CREB (A). RAC1 and CREB co-localization (B). Binding relationship of CREB to miR-15b promoter (C). miR-155-5p transfection-mediated changes in CREB and p-CREB levels (D). miR-155-5p transfection-mediated changes in the transcriptional activity of miR-15b promoter and miR-15b expression (E-F).
|
Overexpression of CREB or miR-15b suppresses miR-155-5p-induced apoptosis of CD34+ cells.
In CD34+ cells transfected with Lv-miR-155-5p mimic + CREB or
Lv-miR-155-5p mimic + miR-15b mimic, the transfection efficacy was
checked to be successful (Figure 6A). Due to CREB or miR-15b overexpression, miR-155-5p upregulation-mediated effects on CD34+ cell proliferation (Figure 6B), cell cycle arrest, apoptosis rates (Figure 6C-E), and levels of Bcl-2 and Bax (Figure 6F) were all blocked.
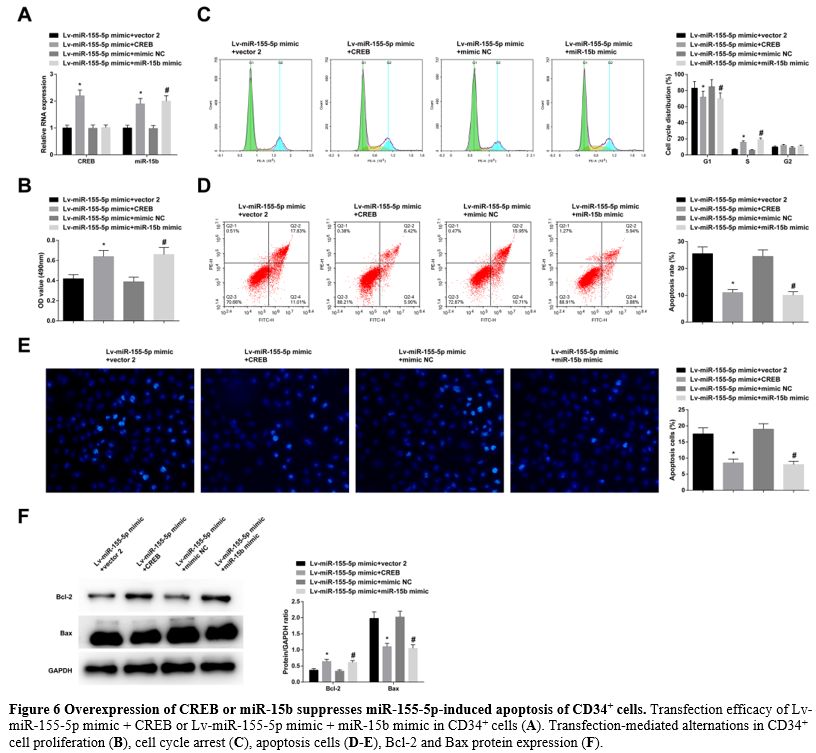 |
- Figure 6 Overexpression of CREB or miR-15b suppresses miR-155-5p-induced apoptosis of CD34+ cells. Transfection efficacy of Lv-miR-155-5p mimic + CREB or Lv-miR-155-5p mimic + miR-15b mimic in CD34+ cells (A). Transfection-mediated alternations in CD34+ cell proliferation (B), cell cycle arrest (C), apoptosis cells (D-E), Bcl-2 and Bax protein expression (F).
|
miR-155-5p promotes PD-L1 expression by RAC1/CREB/miR-15b axis. Immunoblotting for PD-L1 in CD34+
cells found that overexpression of miR-155-5p could promote PD-L1
expression, low expression of miR-155-5p could inhibit PD-L1
expression, while overexpression of RAC1, CREB or miR-15b could prevent
the promoting effect of miR-155-5p on PD-L1 expression (Figure 7A, B).
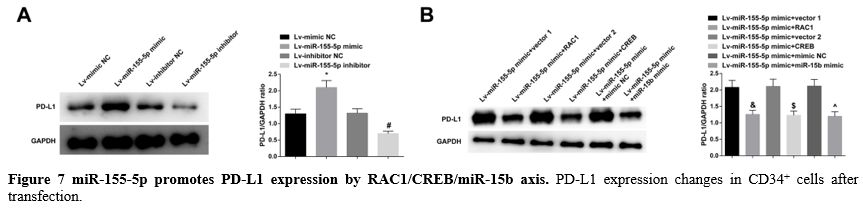 |
- Figure 7 miR-155-5p promotes PD-L1 expression by RAC1/CREB/miR-15b axis. PD-L1 expression changes in CD34+ cells after transfection.
|
Discussion
MDS
consists of a constellation of bone marrow failure diseases and
ineffective hematopoiesis is the main obstacle to disease management.
CD34 family is a marker for type I transmembrane sialomucin and HSCs,
and bioinformatics analysis in CD34+ cells from MDS patients is applicable for evaluating miRNA-mRNA interaction.[19]
To improve MDS, targeting treatment strategies to increase the
effectiveness of hematopoiesis is of great therapeutic potential.[20] Similar to the treatment theory, the study from the aspect of miRNA evaluated the action of miR-155-5p in CD34+ MDS cell activities and eventually summarized that miR-155-5p induces CD34+ MDS cell apoptosis by modulating RAC1-mediated CREB/miR-15b axis and activating PD-L1 checkpoint.
miR-155-5p is one of the best-characterized miRNAs and exerts a key
role in various physiological and pathological processes including
hematopoietic lineage differentiation.[21] In the
course of MDS, miR-155-5p has been validated to present an up-trend
expression level and is associated with high-risk MDS.[5] Furthermore, the increase of miR-155-5p has been also checked in the bone marrow of acquired aplastic anemia patients.[22] miR-155-5p concentration is elevated in the serum of AML patients and is feasible to distinguish AML patients.[23]
Moreover, M-K Chuang et al. have measured that hsa-miR-155-5p is high
in de novo acute myeloid leukemia patients and is an independent
indicator of poor prognosis.[24] The current paper also supported the upward expression pattern of miR-155-5p in MDS, as manifested in CD34+
cells, further validated the relationship between high expression of
miR-155-5p and low survival rates of MDS patients, and confirmed the
correlation between miR-155-5p and clinicopathological variables
including BM blasts percentage, IPSS-R, IPSS-M, and mutated genes per
patient. According to a study report, it is known that miR-155
knockdown can prevent the development of myeloproliferative-like
disease and cytokine induction in RBPJ-/- mice.[25]
Notably, it is previously elaborated that AML inhibits hematopoiesis by
producing exosomes carrying miR-150 and miR-155 to target c-MYB.[26]
With regard to hematopoiesis, this research obtained the findings that
miR-155-5p overexpression reduced proliferation and induced apoptosis
of CD34+
cells, as demonstrated in cell cycle, apoptosis rates, expression of
apoptosis-related proteins, and PD-L1 expression. Oppositely,
miR-155-5p silencing attenuated MDS CD34+ cell apoptosis.
Confirmation was obtained regarding the targeting relationship between
miR-155-5p and RAC1 in the study. Activation of RAC1 promotes AML
development by enhancing homing and retention of leukemia cells in the
niche,[27] and RAC1 inhibitors have suggested an anti-proliferative effect on AML cells.[28]
Additionally, it is reported that blocking RAC1 results in impaired
migration by upregulating a dominant negative form of RAC1 in CD34+ cells from leukemia patients.[29]
Intriguingly, activating RAC1 contributes to the promotion of HSC
regeneration, hematologic recovery, and HSC survival under the
treatment of PTPσ inhibitors.[30] In particular, this study discovered that RAC1 was lowly expressed in MDS CD34+
cells and RAC1 induction was protective to prevent miR-155-5p-induced
cellular apoptosis, thereby enhancing bone marrow hematopoiesis. As
mechanistically determined, miR-155-5p reduced the transcriptional
activity of miR-15b by inhibiting RAC1, dissociating the interaction
between RAC1 and CREB, and inhibiting CREB activation. Also, either
overexpressing CREB or miR-15b reduced miR-155-5p-mediated apoptosis of
CD34+ cells. CREB has been commonly accepted as a pro-oncogene in AML,[31,32]
but its regulatory action in MDS has been rarely reported. It is
established that loss of miR-15/16 can lead to MDS-AML transformation.[14]
Conclusions
The paper illustrates that miR-155-5p mediates PD-L1 and promotes CD34+
cell apoptosis in MDS by regulating the RAC1/CREB/miR-15b axis, thereby
inhibiting bone marrow hematopoiesis. To some extent, the findings may
open up a new avenue for the development of molecule-based
pharmaceutics for MDS. However, the relevant mechanism was only studied
in cells, but not verified in animal experiments. Also, more rigorous
experimental protocols should be designed to validate the role and
mechanism of miR-155-5p in MDS.
Funding
This
work was supported by grants from the National Natural Science
Foundation of China (No. 82000129), Guangdong Basic and Applied Basic
Research Foundation (No. 2019A1515110160), Chinese Traditional Medicine
of Guangzhou Science and Technology Project (No. 20202A011012), Funding
of Guangzhou Institute of Pediatrics/Guangzhou Women and Children’s
Medical Center (No. YIP-2018-050, Pre-NSFC-2019-004, Pre-NSFC-2019-005).
Ethics Approval and Consent to Participate
The
present study was approved by the Ethics Committee of Guangzhou Women
and Children’s Medical Center, Guangzhou Medical University and written
informed consent was provided by all patients prior to the study start.
All procedures were performed in accordance with the ethical standards
of the Institutional Review Board and The Declaration of Helsinki, and
its later amendments or comparable ethical standards.
References
- Cazzola, M., Myelodysplastic Syndromes. N Engl J Med, 2020. 383(14): p. 1358-1374. https://doi.org/10.1056/NEJMra1904794 PMid:32997910
- Saygin,
C. and H.E. Carraway, Current and emerging strategies for management of
myelodysplastic syndromes. Blood Rev, 2021. 48: p. 100791. https://doi.org/10.1016/j.blre.2020.100791 PMid:33423844
- Zhou, J., A. Orazi, and M.B. Czader, Myelodysplastic syndromes. Semin Diagn Pathol, 2011. 28(4): p. 258-72. https://doi.org/10.1053/j.semdp.2011.08.005 PMid:22195404
- Haferlach, T., The Molecular Pathology of Myelodysplastic Syndrome. Pathobiology, 2019. 86(1): p. 24-29. https://doi.org/10.1159/000488712 PMid:29791902
- Wan,
C., et al., Microarray analysis of differentially expressed microRNAs
in myelodysplastic syndromes. Medicine (Baltimore), 2020. 99(27): p.
e20904. https://doi.org/10.1097/MD.0000000000020904 PMid:32629683 PMCid:PMC7337584
- Lee,
D.W., et al., Loss of SHIP-1 protein expression in high-risk
myelodysplastic syndromes is associated with miR-210 and miR-155.
Oncogene, 2012. 31(37): p. 4085-94. https://doi.org/10.1038/onc.2011.579 PMid:22249254
- Cao,
M., et al., Mechanisms of Impaired Neutrophil Migration by MicroRNAs in
Myelodysplastic Syndromes. J Immunol, 2017. 198(5): p. 1887-1899. https://doi.org/10.4049/jimmunol.1600622 PMid:28130497
- Kalfa,
T.A., et al., Rac1 and Rac2 GTPases are necessary for early
erythropoietic expansion in the bone marrow but not in the spleen.
Haematologica, 2010. 95(1): p. 27-35. https://doi.org/10.3324/haematol.2009.006239 PMid:20065081 PMCid:PMC2805739
- Nguyen,
T.M., et al., EphA5 and EphA7 forward signaling enhances human
hematopoietic stem and progenitor cell maintenance, migration, and
adhesion via Rac1 activation. Exp Hematol, 2017. 48: p. 72-78. https://doi.org/10.1016/j.exphem.2016.12.001 PMid:27988259
- Bu,
F., et al., Activation of endothelial ras-related C3 botulinum toxin
substrate 1 (Rac1) improves post-stroke recovery and angiogenesis via
activating Pak1 in mice. Exp Neurol, 2019. 322: p. 113059. https://doi.org/10.1016/j.expneurol.2019.113059 PMid:31499064 PMCid:PMC6864282
- Pardo,
L., et al., CREB Regulates Distinct Adaptive Transcriptional Programs
in Astrocytes and Neurons. Sci Rep, 2017. 7(1): p. 6390. https://doi.org/10.1038/s41598-017-06231-x PMid:28743894 PMCid:PMC5526874
- Arige,
V., et al., Regulation of Monoamine Oxidase B Gene Expression: Key
Roles for Transcription Factors Sp1, Egr1 and CREB, and microRNAs
miR-300 and miR-1224. J Mol Biol, 2019. 431(6): p. 1127-1147. https://doi.org/10.1016/j.jmb.2019.01.042 PMid:30738894
- Zhu,
B., et al., Transcriptional regulation of miR-15b by c-Rel and CREB in
Japanese encephalitis virus infection. Sci Rep, 2016. 6: p. 22581. https://doi.org/10.1038/srep22581 PMid:26931521 PMCid:PMC4773857
- Lovat,
F., et al., Combined loss of function of two different loci of
miR-15/16 drives the pathogenesis of acute myeloid leukemia. Proc Natl
Acad Sci U S A, 2020. 117(22): p. 12332-12340. https://doi.org/10.1073/pnas.2003597117 PMid:32424097 PMCid:PMC7275703
- Greenberg,
P.L., et al., Revised international prognostic scoring system for
myelodysplastic syndromes. Blood, 2012. 120(12): p. 2454-65. https://doi.org/10.1182/blood-2012-03-420489 PMid:22740453 PMCid:PMC4425443
- Nachtkamp, K., et al., Myelodysplastic Syndromes. Dtsch Arztebl Int, 2023. 120(12): p. 203-210.
- Lu,
H.T., et al., miR-424-5p regulates apoptosis and cell proliferation via
targeting Bcl2 in nucleus pulposus cells. Anim Cells Syst (Seoul),
2020. 24(3): p. 136-142. https://doi.org/10.1080/19768354.2020.1775699 PMid:33209193 PMCid:PMC7651851
- Wang,
J., et al., Mutual inhibition between YAP and SRSF1 maintains long
non-coding RNA, Malat1-induced tumourigenesis in liver cancer. Cell
Signal, 2014. 26(5): p. 1048-59. https://doi.org/10.1016/j.cellsig.2014.01.022 PMid:24468535
- Xu,
F., et al., Identification of microRNA-regulated pathways using an
integration of microRNA-mRNA microarray and bioinformatics analysis in
CD34+ cells of myelodysplastic syndromes. Sci Rep, 2016. 6: p. 32232. https://doi.org/10.1038/srep32232 PMid:27571714 PMCid:PMC5004188
- Syed,
K., et al., Novel combinations to improve hematopoiesis in
myelodysplastic syndrome. Stem Cell Res Ther, 2020. 11(1): p. 132. https://doi.org/10.1186/s13287-020-01647-1 PMid:32197634 PMCid:PMC7083030
- Elton,
T.S., et al., Regulation of the MIR155 host gene in physiological and
pathological processes. Gene, 2013. 532(1): p. 1-12. https://doi.org/10.1016/j.gene.2012.12.009 PMid:23246696
- Srivastava,
J., et al., Differential expression of miRNAs and their target genes:
Exploring a new perspective of acquired aplastic anemia pathogenesis.
Int J Lab Hematol, 2020. 42(5): p. 501-509. https://doi.org/10.1111/ijlh.13245 PMid:32490599
- Zhi,
F., et al., Identification of circulating microRNAs as potential
biomarkers for detecting acute myeloid leukemia. PLoS One, 2013. 8(2):
p. e56718. https://doi.org/10.1371/journal.pone.0056718 PMid:23437222 PMCid:PMC3577716
- Chuang,
M.K., et al., A 3-microRNA scoring system for prognostication in de
novo acute myeloid leukemia patients. Leukemia, 2015. 29(5): p. 1051-9.
https://doi.org/10.1038/leu.2014.333 PMid:25428263
- Wang,
L., et al., Notch-dependent repression of miR-155 in the bone marrow
niche regulates hematopoiesis in an NF-kappaB-dependent manner. Cell
Stem Cell, 2014. 15(1): p. 51-65. https://doi.org/10.1016/j.stem.2014.04.021 PMid:24996169 PMCid:PMC4398997
- Hornick,
N.I., et al., AML suppresses hematopoiesis by releasing exosomes that
contain microRNAs targeting c-MYB. Sci Signal, 2016. 9(444): p. ra88. https://doi.org/10.1126/scisignal.aaf2797 PMid:27601730
- Chen,
S., et al., Rac1 GTPase Promotes Interaction of Hematopoietic
Stem/Progenitor Cell with Niche and Participates in Leukemia Initiation
and Maintenance in Mouse. Stem Cells, 2016. 34(7): p. 1730-41. https://doi.org/10.1002/stem.2348 PMid:26946078
- Hemsing,
A.L., et al., NPM1-Mutated Patient-Derived AML Cells Are More
Vulnerable to Rac1 Inhibition. Biomedicines, 2022. 10(8). https://doi.org/10.3390/biomedicines10081881 PMid:36009428 PMCid:PMC9405324
- Wang,
J.Y., et al., Activation of Rac1 GTPase promotes leukemia cell
chemotherapy resistance, quiescence and niche interaction. Mol Oncol,
2013. 7(5): p. 907-16. https://doi.org/10.1016/j.molonc.2013.05.001 PMid:23726395 PMCid:PMC5528460
- Zhang, Y., et al., PTPsigma inhibitors promote hematopoietic stem cell regeneration. Nat Commun, 2019. 10(1): p. 3667. https://doi.org/10.1038/s41467-019-11490-5 PMid:31413255 PMCid:PMC6694155
- Tregnago, C., et al., CREB engages C/EBPdelta to initiate leukemogenesis. Leukemia, 2016. 30(9): p. 1887-96. https://doi.org/10.1038/leu.2016.98 PMid:27118402
- Cho, E.C., B. Mitton, and K.M. Sakamoto, CREB and leukemogenesis. Crit Rev Oncog, 2011. 16(1-2): p. 37-46. https://doi.org/10.1615/CritRevOncog.v16.i1-2.50 PMid:22150306 PMCid:PMC3243968
Supplementary material
 |
Supplementary table 1. Chromosome abnormalities in MDS |
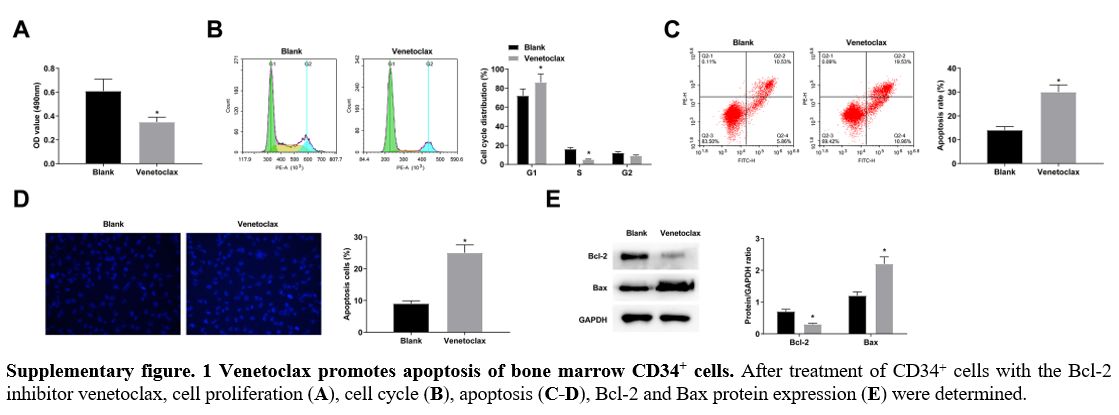 |
Supplementary figure. 1
Venetoclax promotes apoptosis of bone marrow CD34+ cells. After
treatment of CD34+ cells with the Bcl-2 inhibitor venetoclax, cell
proliferation (A), cell cycle (B), apoptosis (C-D), Bcl-2 and Bax protein expression (E) were determined.
|
[TOP]











