Although this variability is not fully understood yet, specific haplotypes around the β-hemoglobin S cluster (commonly referred to as ‘βs-globin haplotypes’ or ‘SCA haplotypes’) are included among genetic factors explaining the phenotypic heterogeneity observed in SCA.[2] Traditionally, βS-globin haplotypes are defined by restriction fragment length polymorphisms (RFLP) using a combination of polymorphic restriction sites in a genomic region of ~60kb located within the HBB cluster on chromosome 11 and are inherited along with the SCA mutation. The five classical SCA haplotypes are named according to the geographical region where they were originally identified. Apart from them, there are some unusual, less frequent haplotypes known as atypical βS haplotypes.[3]
Modifying effects of SCA haplotypes are in part related to their association with the level of fetal hemoglobin (HbF), which is considered the most powerful modulator of the clinical expression of SCA.[4] The Central African Republic (CAR) haplotype is known to present a severe phenotype due to its association with a low average HbF level of about 5%. In comparison, the Cameroon (CAM) and the Benin (BEN) haplotypes, with an intermediate level of HbF of 7%, have moderate symptomatology. The Senegal (SEN) and Arabian Indian (AI) haplotypes, which are associated with 10 to 20% HbF in adult SCA patients, present milder phenotypic patterns.[5]
Therefore, each haplotype, with its related phenotype severity, has predictive value in the management of SCA, allowing personalized care.[6]
The Democratic Republic of Congo (DRC) is among the most affected countries by SCA worldwide, with an incidence of around 2% in newborns.[7] Still, data on the effect of SCA haplotypes on disease severity are scarce for the DRC. Yet this knowledge could be useful to explain the phenotypical diversity previously observed in Congolese SCA patients.[8]
The distribution of SCA haplotypes, as well as their correlation with baseline hematological parameters, were assessed in a cohort of SCA patients in the DRC.
Material and Methods
Patient selection. This was a cross-sectional study conducted at Kisantu St Luc Hospital (KSLH), located in the Kongo Central Province, and at the “Centre de Médecine Mixte et d’Anémie SS” (CMMASS), in Kinshasa. Pediatric patients were recruited at KSLH, whereas adult patients were recruited at "CMMASS Recruitment." The study was performed during regular SCA follow-ups at both sites. Prior to the inclusion in this study, a SCA diagnosis resulting from the homozygous presence of the p. Glu7Val mutation in HBB was confirmed by molecular testing, as previously described.[9] In addition, we excluded, among confirmed SCA patients, those not in a steady state of the disease, those who received a transfusion within 4 months before blood sample collection, and those treated with hydroxyurea.Sample collection. For each participant, peripheral blood for hematologic tests and DNA extraction was collected in two 4 ml EDTA-coated tubes.
Hematological variables. A blood count was performed on an automated counter (Sysmex Hematology Counter, Japan). The measured hematologic parameters were hemoglobin (Hb), white blood cells (WBC), platelets (PLT), and reticulocytes (RETIC). The HbF level was quantified by capillary electrophoresis (Minicap, SEBIA, France).
DNA extraction and quality control. Genomic DNA was extracted from blood samples using the salting out method[10] and stored at -20°C at the Center for Human Genetics of the University of Kinshasa (DR Congo). The concentration and purity of the extracted DNA were evaluated using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, USA).
SNP genotyping and processing. Our single nucleotide polymorphisms (SNPs) set contained the 4 polymorphisms defining the five βs-globin haplotypes.[11] An additional 63 SNPS were included as part of this larger study of other genomic modifiers and pharmacogenomic variants.
Molecular Inversion Probes (MIPs) 108 to 120 bp long were ordered from NimaGen (Netherlands). The design's genomic coordinates were based on GRCh37.
Samples were normalized to 100 ng/µl at the Genomics Core at the Center for Human Genetics of the University Hospitals Leuven (UZ Leuven) and checked on a Qubit 2.0 Fluorometer (Life Technologies, Bleiswijk, Netherlands). Patient-only targeted sequencing was performed using the Single-Stranded Molecular Inversion Probe (ssMIP) approach, using a custom design probe set and the standard ssMIP procedure.[12]
A total of 100 ng DNA was used for library preparation. After capturing with ssMIPs, samples were pooled by 76 for pools 1 and 2, by 86 for pool 3, and by 100 for pool 4. Pool 4 contained both the remaining samples and samples that failed in the other three pools. To reach our target coverage of 500x, pools 1, 2, and 3 were sequenced on a MiSeq v3 PE300 cycles flow cell (13.2–15 Gb output), whereas pool four was run on a MiSeq v2 PE300 cycles (4.5–5.1 Gb output). A concentration of 12.5pM was loaded on the MiSeq, as per the recommendations in the MiSeq manual (Illumina, San Diego, California, USA). A PhiX spike-in of 3% was also added. The quality control of sequencing reads was performed using FastQC. Overlapping paired-end reads from the same fragment were merged using FLASH 1.2.11.[13] Reads were then mapped against the reference genome build GRCh37 with BWA-mem 0.7.17.[14] Duplicate reads were kept. All positions were genotyped with GATK HaplotypeCaller 4.0.11.0 with the option EMIT_ALL_SITES.[15] All samples were called together using GATK GenotypeGVCFs 3.8-0.
Data analysis. Quantitative data of white blood cells, platelet, and reticulocyte counts were normalized by log10-transformation prior to statistical analysis. For each of the outcomes, a regression analysis was performed for the SNP genotype using an additive model and with age and sex as covariates using PLINK v1.9 (https://www.cog-genomics.org/plink/1.9). An uncorrected p-value ≤ 0.05 was considered significant.
The SCA haplotypes were manually derived based on the genotype of the 4 SNPs of interest.
Ethics statement. Written informed consent was obtained from adult patients or from parents of children. The study protocol was approved by the Ethical Committee of the Public Health School at the University of Kinshasa, DRC (Protocol number ESP/CE/079/2016).
Results
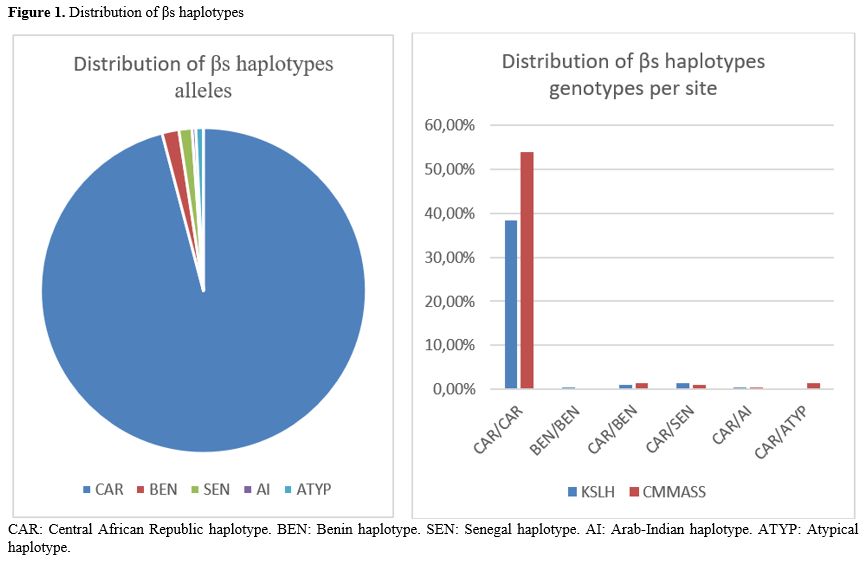
Description of the cohort. A total of 277 SCA patients were included, aged between 2 and 40 years. The sex distribution was 112 (40%) males and 165 (60%) females. After removing eight samples with less than 85% of SNPs correctly genotyped, samples of 269 patients were considered: 112 patients (42%) from KSLH, including 54 males and 58 females aged between 2 and 17 years, and 157 patients (58%) from CMMASS, comprising 56 males and 101 females aged between 18 and 40 years.Distribution of SCA haplotypes. Among the 269 patients (538 alleles), the CAR haplotype was the major allele in this study, with 95.9% (516/538). The other haplotypes were BEN 1.6% (9/538), SEN 1.3% (7/538), and AI 0.3% (2/538). When grouped at genotype levels (i.e., the composition of the two haplotype alleles for a particular SNP), a total of 5 different genotypes were identified, including homozygous CAR 92.2% (248/269), homozygous BEN 0.3% (1/269), and double heterozygous CAR/BEN 2.5% (7/269), CAR/SEN 2.5% (7/269) and CAR/AI 0.7% (2/269) (Figure 1).
Three different atypical haplotypes were identified. Atypical haplotypes accounted for 0.7 % (4/538) of the alleles. Genotypically, all patients with atypical alleles were heterozygous CAR/ATYP (1.4%, 4/269). The atypical profiles were confirmed by Sanger sequencing to exclude the possibility of a false call.
The profile and frequency of classical and atypical haplotype alleles are presented in Table 1.
Influence of SCA haplotypes on hematological parameters. The almost homogeneous distribution of SCA haplotypes in our cohort, with a prevalence of over 92% of CAR homozygotes, did not allow evaluation of the influence of haplotypes on hematological parameters.
Discussion
This study assessed the influence of SCA haplotypes as modulators of the biological expression of SCA in a cohort of Congolese SCA patients.A significant predominance of the CAR (Central African Republic) haplotype was observed at a frequency higher than 95%. The same trend has been recently reported in a multicentric study involving patients from DRC and other neighboring African countries, including Angola, Uganda, and Kenya (CAR-CAR at 92% in 635 SCA children).[16] The homogeneity of the βS haplotype in the regions we studied may reflect limited admixture with other populations.
This observation of a nearly homogeneous haplotype genotype contrasts with studies from other African, European, and American countries. In many American countries, the heterogeneity of βS haplotypes is explained by the flow of Africans from different regions during the slave trade period. In Europe, the diversity of βS haplotypes is essentially due to migrations from several African regions. In Africa, the distribution of βS haplotypes is highly diversified. In some countries, such as Cameroun and Egypt, all βS haplotypes are represented, albeit with a higher frequency of the Benin haplotype and a lower frequency of the Arab-Indian haplotype.[3]
However, the presence of the BEN, SEN, and AI haplotypes in our cohort, even at very low frequency, could result from the recent admixture of West Africans, Portuguese, Lebanese, and Indians with the local population.[17] We could not find any anthropological explanation for the absence of the CAM haplotype in our study sample.
The frequency of 0.74% of atypical haplotypes reported in our study is lower than the 5 to 10% often observed concerning the whole βS locus.[18] Population's mixture is among the factors involved in the multiplicity of atypical βS haplotypes. Therefore, our finding can be explained by the relative homogeneity of the studied population.
The predominance of one specific SCA haplotype and very low frequencies of others did not allow proper assessment of the influence of haplotypes on the variability of hematological profile in our study cohort. Bitoungi reached a similar conclusion in a study involving one of the largest national cohorts of sub-Saharan African SCA patients, where the authors found mainly the BEN haplotype.[19] Overall, the lack of haplotype diversity in those African populations prevents the evaluation of haplotypes' contribution among patients sharing the same environment.
Traditionally, the βS-globin haplotypes are typically characterized using a combination of four to eight restriction enzymes, each recognizing specific restriction sites. However, the RFLP method is not only time-consuming but also may result in unusual restriction patterns in the presence of unexpected SNP at the restriction site without precisely determining the nucleotide change at the polymorphic site.[20] Nowadays, the application of Next Generation Sequencing (NGS) techniques as the MIP method offers advantages by its ability to multiplex samples and interrogate multiple loci in a single experiment and the possibility of fully characterizing atypical βs haplotypes.[11]
Conclusions
The CAR haplotype is predominant among DR Congolese SCA patients. The evaluation of the influence of the βs haplotypes on hematological parameters is limited by the very low representativeness of other haplotypes. Therefore, this study highlights that phenotypical diversity observed in Congolese SCA patients is not related to βS-globin haplotypes. Other factors involved need to be explored.References
- Aghajani F, Mahdavi MR, Kosaryan M, Mahdavi M,
Hamidi M, Jalali H. Identification of β-globin haplotypes linked to
sickle hemoglobin (HbS) alleles in Mazandaran province, Iran. Genes
Genet Syst.2017; 91(6):311-313 https://doi.org/10.1266/ggs.16-00005 PMid:28003571
- Ndugwa
C, Higgs D, Fisher C, Hambleton I, Mason K, Serjeant BE, Serjeant GR.
Homozygous sickle cell disease in Uganda and Jamaica a comparison of
Bantu and Benin haplotypes. West Indian Med J. 2012; 61(7):
684-91
- Kevin
Esoh & Ambroise Wonkam, Evolutionary history of sickle-cell
mutation: implications for global genetic medicine, Human Molecular
Genetics, Volume 30, Issue R1, 1 March 2021, Pages R119-R128, https://doi.org/10.1093/hmg/ddab004 PMid:33461216 PMCid:PMC8117455
- Al-Saqladi
AW, Brabin BJ, Bin-Gadeem HA, et al. Beta-globin gene cluster
haplotypes in Yemeni children with sickle cell disease. Acta Haematol.
2010 ;123(3) :182-185 https://doi.org/10.1159/000294965 PMid:20224271
- Bhagat
S, Patra PK, Thakur AS. Fetal Haemoglobin and β-globin Gene Cluster
Haplotypes among Sickle Cell Patients in Chhattisgarh. J Clin Diagn
Res. 2013 Feb;7(2):269-72 https://doi.org/10.7860/JCDR/2013/4381.2744 PMid:23542314 PMCid:PMC3592290
- Adekile
A, Akbulut-Jeradi N, Al Khaldi R, et al. Diagnosis of Sickle Cell
Disease and HBB Haplotyping in the Era of Personalized Medicine: Role
of Next Generation Sequencing. J Pers Med. 2021 ;11(6) :454 https://doi.org/10.3390/jpm11060454 PMid:34071035 PMCid:PMC8224627
- Tshilolo
L, Aissi LM, Lukusa D, Kinsiama C, Wembonyama S, Gulbis B, Vertongen F.
Neonatal screening for sickle cell anaemia in the Democratic Republic
of the Congo: experience from a pioneer project on 31 204 newborns. J
Clin Pathol.2009; 62(1) : 35-8 https://doi.org/10.1136/jcp.2008.058958 PMid:19103857
- Mikobi
TM, Lukusa Tshilobo P, Aloni MN, et al. Clinical phenotypes and the
biological parameters of Congolese patients suffering from sickle cell
anemia: A first report from Central Africa. J Clin Lab Anal. 2017
;31(6): e22140 https://doi.org/10.1002/jcla.22140 PMid:28116772 PMCid:PMC6816843
- Ngole
M, Race V, Mbayabo G, Lumbala P, Songo C, Lukusa PT, Devriendt K,
Matthijs G, Lumaka A . DNA testing for sickle cell anemia in Africa:
Implementation choices for the Democratic Republic of Congo. J Clin Lab
Anal. 2022;36(5):e24398 https://doi.org/10.1002/jcla.24398 PMid:35405024 PMCid:PMC9102645
- Miller
SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting
DNA from human nucleated cells. Nucleic Acids Res.1988;16(3) :1215 https://doi.org/10.1093/nar/16.3.1215 PMid:3344216 PMCid:PMC334765
- Shaikho
EM, Farrell JJ, Alsultan A, Qutub H, Al-Ali AK, Figueiredo MS, Chui
DHK, Farrer LA, Murphy GJ, Mostoslavsky G, Sebastiani P, Steinberg MH .
A phased SNP-based classification of sickle cell anemia HBB haplotypes.
BMC Genomics.2017;18(1):608 https://doi.org/10.1186/s12864-017-4013-y PMid:28800727 PMCid:PMC5553663
- Cantsilieris
S, Stessman HA, Shendure J, Eichler EE. Targeted Capture and High
Throughput Sequencing Using Molecular Inversion Probes (MIPs). Methods
Mol Bio.2017;1492:95- 106 https://doi.org/10.1007/978-1-4939-6442-0_6 PMid:27822858 PMCid:PMC5484527
- Magoč
T, Salzberg SL. FLASH: fast length adjustment of short reads to improve
genome assemblies. Bioinformatics.2011;27(21):2957-63 https://doi.org/10.1093/bioinformatics/btr507 PMid:21903629 PMCid:PMC3198573
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14), 1754-176 https://doi.org/10.1093/bioinformatics/btp324 PMid:19451168 PMCid:PMC2705234
- McKenna
A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella
K, Altshuler D, Gabriel S, Daly M, DePristo MA . The Genome Analysis
Toolkit: a MapReduce framework for analyzing next-generation DNA
sequencing data. Genome Res.2010;20(9),1297-303 https://doi.org/10.1101/gr.107524.110 PMid:20644199 PMCid:PMC2928508
- Tshilolo
L, Tomlinson G, Williams TN, Santos B, Olupot-Olupot P, Lane A, Aygun
B, Stuber SE, Latham TS, McGann PT, Ware RE; REACH Investigators.
Hydroxyurea for Children with Sickle Cell Anemia in Sub-Saharan Africa.
N Engl J Med.2019;380(2) :121-131 https://doi.org/10.1056/NEJMoa1813598 PMid:30501550 PMCid:PMC6454575
- Flahaux
ML, Schoumaker B. Democratic Republic of the Congo: A Migration History
Marked by Crises and Restrictions. The online Journal of Migration
Policy Institute;2016: April 20. www.migrationpolicy.org. Accessed March 15, 2022
- Zago
MA, Silva WA Jr, Dalle B, et al. Atypical beta(s) haplotypes are
generated by diverse genetic mechanisms. Am J Hematol. 2000 ;63(2)
:79-84 https://doi.org/10.1002/(SICI)1096-8652(200002)63:2<79::AID-AJH4>3.0.CO;2-D
- Bitoungui
VJ, Pule GD, Hanchard N, Ngogang J, Wonkam A. Beta-globin gene
haplotypes among cameroonians and review of the global distribution: is
there a case for a single sickle mutation origin in Africa?
OMICS.2015;19(3):171-9 https://doi.org/10.1089/omi.2014.0134 PMid:25748438 PMCid:PMC4356477
- Joly
P, Lacan P, Garcia C, et al. Rapid and reliable β-globin gene cluster
haplotyping of sickle cell disease patients by FRET Light Cycler and
HRM assays. Clin Chim Acta. 2011 ;412(13-14) :1257-1261 https://doi.org/10.1016/j.cca.2011.03.025 PMid:21440534