Multiparametric flow cytometry (MCF) is a well-established and user-friendly single-cell technology that simultaneously measures multiple analyte expression patterns in individual cells.
Immunophenotype characterization performed by MFC is an essential step for ALL diagnosis and has significant relevance in the evaluation of minimal residual disease (MRD).[2] Indeed, leukemic cells express surface and intracytoplasmic antigens whose identification allows us to determine the line of belonging, the level of differentiation and maturation, the lineage infidelity, and peculiar aberrations.[3,4] In particular, B-ALLs EGIL classification (European Group for the immunological classification of leukemias),[5-6] in addition to the expression of B lineage antigens, is based on the detection of cytoplasmic IgM (cIgM) and CD10 on leukemic B cells. The expression of TdT/CD19/CD22/cCD79a with or without CD20/CD34, in the absence of cIgM and CD10, identifies the pro-B ALL subtype. The presence of CD10 antigen (CALLA) without cIgM defines the B-common ALL, while CD10+/– expression associated with cIgM identifies pre-B ALL. Finally, the presence of surface Ig light chains defines mature B-ALL.[7] The most important and prognostically and therapeutically relevant distinction for B-ALL is between Ph-positive (Ph+) and Ph-negative (Ph-) B-ALL. Ph+ ALLs are defined by the presence of the Philadelphia chromosome (Ph) generated by the translocation between chromosomes 9 and 22, with t(9;22)(q34;q11) being the most common chromosomal abnormality in adults.[8-10]
This aberration produces the BCR::ABL1 fusion gene, which encodes for a constitutively activated tyrosine kinase signaling protein that sustains leukemic cell genesis and proliferation.[8] Ph chromosome has an overall incidence of roughly 20% to 25% in B-ALLs.[11-12] This translocation was considered one of the worst prognostic factors before the introduction of tyrosine kinase inhibitors (TKIs) in clinical practice.[13]
TKIs revolutionized the outcome of ALL-Ph+ patients, with chemotherapy and allogeneic stem cell transplant (allo-HSCT) questioned by the usage of chemo-free approaches.[14]
In 2009, the term "Philadelphia–like" or "BCR::ABL1–like" ALL was first used.14 Ph-like ALLs are characterized by a gene expression profile (GEP) highly similar to that of BCR::ABL1-positive ALL but lacking the BCR::ABL1 fusion protein derived from the t(9;22)(q34;q11) translocation, with high frequency of deletions of IKZF1 gene, encoding the lymphoid transcription factor IKAROS, and other lymphoid transcription factor genes, such as PAX5 and EBF1.[15-19]
Ph-like ALLs also have a phenotypic expression profile similar to Ph+ ALL, presenting myeloid antigens such as CD13/CD33 and low expression of CD38. Importantly, Ph-like ALLs have a poor outcome, and therefore, their distinction from the other subgroups of ALL is fundamental.
Cluster of differentiation 146 (CD146, also known as Mel-CAM or MUC18) is a cell transmembrane glycoprotein belonging to the immunoglobulin family and represents an adhesion molecule first discovered on the plasma membrane of human melanoma cells, and it was initially named MCAM (melanoma cell adhesion molecule).[20] This 113-kDa glycoprotein is expressed in normal tissues, including smooth muscles, mesenchymal cells, and vascular endothelium on the entire vascular tree that exerts cation-independent adhesion through interactions with an unidentified ligand.[21]
CD146 presents multifunctional activities in both physiological and pathological conditions, including immunity, angiogenesis, and development. CD146 is also expressed in several cancers. A growing number of studies suggest that CD146 overexpression was significantly correlated with the progression, angiogenesis, and metastasis of different malignant tumors and, especially for solid neoplasms, was associated with poor survival and might be considered as a useful prognostic biomarker and promising therapeutic target.[22] The role of CD146 in hematopoietic cells has yet to be thoroughly understood, and few data are available: CD146 is rarely expressed in acute myeloid leukemia (AML) cells, while it can be found frequently in ALLs. In particular, an Italian study showed that only 3.3% of AML were CD146 positive, and these cases were indeed classified as AML, which was not otherwise specified. Conversely, 66% of T-ALLs and 36.8% of B-ALLs, comprising Ph+ ALL cases, expressed CD146 on the blast cells.[23] Hence, CD146 antibody inclusion in MFC panels for suspected acute leukemia (AL) may improve accurate diagnostic workup.
Therefore, the purpose of our study is to propose an optimized MFC diagnostic panel of routine antigens to predict the presence of BCR quickly: ABL1 rearrangement or Ph-like ALL, by evaluating CD146 expression as a possible marker that may aid in the prompt identification of these specific subgroups of B-ALLs.
Materials and Methods
criteria.5-6 Median age was 56 years (range 17-90) years. Patients’ bone marrow (BM) samples were obtained with informed consent in accordance with the Declaration of Helsinki.
B-ALL diagnosis by MCF analysis. B-ALL diagnosis was assessed by MFC using a combination of monoclonal antibodies (mAbs) recommended by the EuroFlow Consortium.[26] BM cells (0.5 × 106) were first stained using a combination of mAbs directed against myeloid (MPO), B and T lymphoid (cCD79a and cCD3) lineage antigens; after that, with a combination of mAbs against:
CD45/CD10/CD34/TdT/HLADR/CD19/CD22/CD20/CD38/CD58/CD123/Igκ/Igλ/cIgM/CD3/CD13/CD33/CD66c/NG2 (Becton Dickinson, San Jose, CA; Società Italiana Chimici, SIC, Life Sciences, Rome, Italy, Beckman Coulter, Brea, CA).
In all B-ALL samples, CD146 (Società Italiana Chimici, SIC, Life Sciences, Rome, Italy) analysis was optimized using additional combinations of monoclonal antibodies as follows:
CD58FITC/CD146PE/CD19PECy7/CD34APC/CD20APC-Cy7/CD13R718/CD38V450/CD45V500/CD10 BV605/CD33 BV711/CD22 BV786).
Data on standardized 12 color staining combinations were acquired on FACSLyric flow cytometers (Becton Dickinson) by collecting at least 50,000 ungated events and analyzed using the PAINT-A-GATE and FACSDIVA softwares (Becton Dickinson). Cytometer setup and tracking beads (BD) were used for daily cytometer optimization. Leukemic cells were gated within the total CD45+ leukocyte population, considering that all cases of B-lineage ALLs were positive for the pan-leukocyte antigen. Representative plots of the flow gating strategy are reported in Figure 1.
The presence of pathological cells was identified in comparison with the known patterns of antigen expression by normal maturing lymphoid precursors and was quantified as a percentage of total leukocytes. In all cases, antigen expression was defined by the percentage of blast cells that resulted in positive for the different markers in the immunological gate.[5,27] CD146 expression was assessed on the blast immunological gate. Cell surface antigen expression was quantified on the same flow cytometer and with the same mAbs combination as the mean fluorescent intensity (MFI) of values obtained with specific mAbs compared with values given by the isotype controls. A sample was considered positive for surface antigens if ≥20% of leukemic cells exhibited fluorescence compared with negative control. A threshold ≥10% of gated blasts was used to define the positivity of CD146 expression based on the results of the Receiver Operating Characteristic (ROC) curve for discrimination between ALL-Ph+ and ALL-Ph negative.
Molecular testing. Molecular analysis of BM samples was carried out using a nested approach using a Multiplex RT-PCR system. As previously described,[28] the screening with Multiplex-RT-PCR was designed to detect simultaneously and in a quick time the most common fusion genes in T-ALL rather than in B-ALL: TCF3::PBX1, ETV6::RUNX1, SIL::TAL1, NUP98::RAP1GDS1, SET::NUP214, BCR::ABL1 p190 (e1a2) and p210 (e13a2, e14a2), KMT2A::AFF1 and KMT2A::MLLT1, with two genes screened, KMT2A::AFF1 and KMT2A::MLLT1.
Statistical methods. Summary statistics (mean and standard deviation, median, and range) were reported by category groups. Mann-Whitney or Kruskal-Wallis test for independent groups was used to compare categories. Differences in the study groups were estimated using the chi-square test or the Fisher exact test for categorical covariate. The optimal cut-off for CD146 expression was identified as the optimal threshold through ROC curve analysis. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), associated with the 10% cut-off of CD146, were reported in supplementary figure 1. A multivariate logistic regression model was used to evaluate the independent role of biomarkers. All tests were two-sided, accepting p<0.05 as indicating a statistically significant difference. All analyses were performed using R software.
Results
Within the 79 B-neg-ALLs, 62 (79%) were phenotypically classified as B-common ALL, 12 (15%) as pro-B, 3 (4%) as pre-B, and 2 (2%) as B-mature. The mean blast percentage was 66%±23% (range 20-96), characterized by a mean percentage of CD146 expression of 15%±29% (range 0-90), with a mean MFI of 32 ± 80 (range 0-550) on leukemic cells.
Out of 144 Ph+ ALL cases, 135 were phenotypically subclassified cases (94%) as B-common ALL, 7 as pre-B (5%), and 2 as pro-B (1%). The mean blast percentage was found to be 67%±22% (range 23-97). In this subcategory, the mean percentage of CD146 expression on blast cells was 29%±34% (range 0-97) with a mean MFI of 89±136 (range 1-548).
Within the 19 Ph-like ALL cases, all B-common ALL (100%) were identified.23 The mean blast percentage was 75%±20% (range 26-90). Ph-like leukemic B cells were characterized by a mean percentage of CD146 expression of 11%±28% (range 0-88) with a mean MFI of 48±144 (range 2-620).
Considering the total of 98 Ph-neg ALL cases, the mean blast percentage was 68%±23% (range 20-96), characterized by a mean percentage of CD146 expression of 14%±28% (range 0-90), with a mean MFI of 36 ± 95 (range 0-620) on leukemic cells.
CD146 differential antigen detection and surface expression intensity. We evaluated the differential expression of various CDs in the 3 groups to identify the potential association between CD146 expression and its MFI with one or more B-ALL subtypes. By ROC analysis, a value of 10% was pinpointed as the optimal cut-off in CD146 expression to maximize the separation between the BCR::ABL1-positive and BCR::ABL1-negative groups (Supplementary Figure 1). Therefore, CD146 was considered positive for surface expression if ≥10% of leukemic cells exhibited fluorescence compared with negative control, while a classical threshold of ≥20% was considered for the other surface antigens.
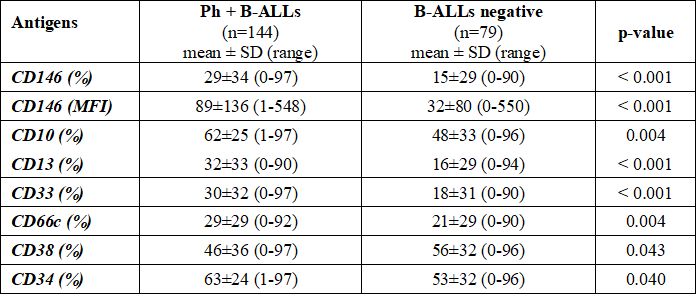
Firstly, we compared Ph+ ALL with B-neg ALL. As expected and previously described in many studies,[29-30] Ph+ B-ALLs were characterized by higher mean expression of myeloid markers such as CD13 (32%±33% vs. 16%±29%, p<0.001), CD33 (30%±32% vs 18%±31%, p<0.001), CD66c (29%±29% vs 21%±29%, p=0.004) and by lower mean expression of CD38 (46%±36% vs 56±32%, p=0.043) (Table 1). Such data was confirmed by higher positivity detection rate for CD13 [74/144 (51%) vs 16/79 (20%), p<0.001], CD33 [72/144 (50%) vs. 22/79 (28%), p=0.002], CD66c [74/144 (51%) vs. 28/79 (35%), p=0.025] and lower number of cases positive for CD38 [94/144 (65%) vs 63/79 (80%), p=0.024] (Supplementary Table 1). Moreover, due to the higher percentage of B-common ALL in the Ph+ group, CD10 presented a higher mean expression (62%±25% vs. 48%±33%, p=0.004) and was more expressed in such group [133/144 (92%) vs. 57/79 (72%), p<0.001]. As for CD146, Ph+ ALLs showed a higher mean expression (29%±34% vs 15%±29%, p<0.001) as well as MIF (89±136 vs 32±80, p<0.001), and greater positivity detection rate [74/144 (51%) vs. 22/79 (28%), p<0.001] of this antigen when compared with B-neg ALLs (Table 1 and Supplementary Table 1).
 |
|
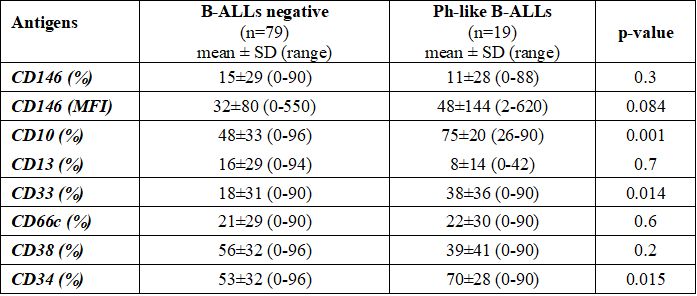
Between Ph-like ALL and B-neg ALL, statistically significant differences in the mean expression of CD33 myeloid marker (38%±36% vs 18%±31%, p=0.014) and in its positivity rate [12/19 (63%) vs 22/79 (28%), p=0.004] were found. CD38 had a lower expression rate in Ph-like ALL cases compared to B-neg ALL cases [10/19 (53%) vs 63/79 (80%), p=0.021]. Due to the presence of only B-common ALL cases in the Ph-like group CD10 resulted in a higher mean expression (75%±20% vs. 48%±33%, p=0.001) and more detected [19/19 (100%) vs. 57/79 (72%), p=0.006] in this group. At variance, no difference was found in CD13 and CD66c expression (p=0.7 and p=0.6, respectively); similarly, neither the mean percentage of CD146 expression (11%±28% vs 15%±29%, p=0.3), neither CD146 MFI (48±144 vs 32±80, p=0.084) or CD146 positivity rate [3/19 (16%) vs 22/79 (28%), p=0.39] were statically different between these two groups (Table 2 and Supplementary Table 2).
 |
|
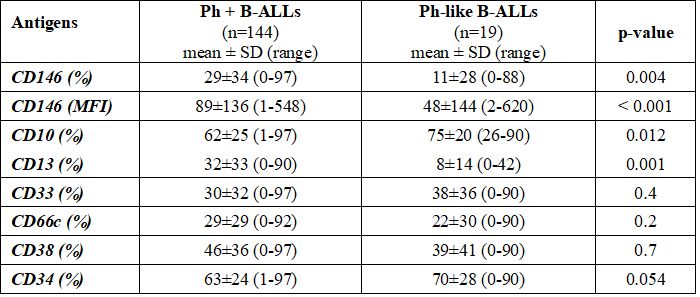
Comparing Ph+ ALLs and Ph-like ALLs, CD33, and CD38 expressions were not found to be statistically significant (p=0.4 and p=0.7, respectively). Indeed, Ph+ blasts cells were characterized by a higher CD13 mean expression (32%±33% vs 8%±14%, p=0.001) and positivity rate [74/144 (51%) vs. 3/19 (16%), p=0.004] as well as by higher CD66c positivity rate [74/144 (51%) vs 5/19 (26%), p=0.043]. Conversely, due to a higher percentage of B-common ALL present in the Ph-like group, CD10 resulted in meanly more expressed in such group (62%±25% vs. 75%±20%, p=0.012). Statistically significant differences emerged in CD146 mean expression between Ph+ and Ph-like B-ALLs (29%±34% vs 11%±28%, p=0.004) as well as in the CD146 MFI (89±136 vs 48±144, p<0.001) and in CD146 positivity rate [74/144 (51%) vs 3/19 (16%), p=0.003] (Table 3 and Supplementary Table 3).
An overall comparison between antigens mean expressions in the three groups is shown in supplementary Table 4, with Ph+ ALLs characterized by a superior CD13, CD33, and CD66c expression, by lower CD38 expression along with the higher mean percentage of CD146 expression and higher CD146 MFI.
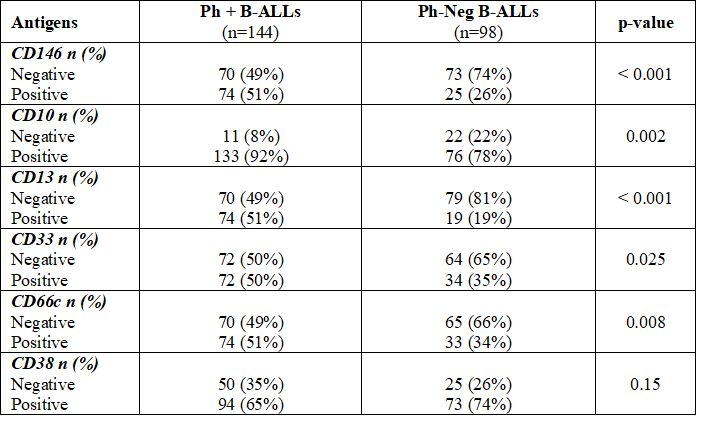
Confronting Ph+ ALLs with all cases of Ph-neg ALLs (B-neg ALL + Ph-like ALLs) a superior mean expression of myeloid markers such as CD13 (32%±33% vs. 14%±27%, p<0.001), CD33 (30%±32% vs 22%±32%, p=0.007) and CD66c (29%±29% vs 22%±29%, p=0.003) was found (Table 4). Such data was confirmed by a higher positivity detection rate for CD13 [74/144 (51%) vs 19/98 (19%), p<0.001], CD33 [72/144 (50%) vs. 34/98 (35%), p=0.025], CD66c [74/144 (51%) vs 33/98 (34%), p=0.008] (Table 5). As for CD146, Ph+ An overall comparison between antigens mean expressions in the three groups is shown in supplementary Table 4, with Ph+ ALLs characterized by a superior CD13, CD33, and CD66c expression, by lower CD38 expression along with the higher mean percentage of CD146 expression and higher CD146 MFI.
Confronting Ph+ ALLs with all cases of Ph-neg ALLs (B-neg ALL + Ph-like ALLs) a superior mean expression of myeloid markers such as CD13 (32%±33% vs. 14%±27%, p<0.001), CD33 (30%±32% vs 22%±32%, p=0.007) and CD66c (29%±29% vs 22%±29%, p=0.003) was found (Table 4). Such data was confirmed by a higher positivity detection rate for CD13 [74/144 (51%) vs 19/98 (19%), p<0.001], CD33 [72/144 (50%) vs. 34/98 (35%), p=0.025], CD66c [74/144 (51%) vs 33/98 (34%), p=0.008] (Table 5). As for CD146, Ph+ ALLs showed a higher CD146 mean expression (29%±34% vs 14%±28%, p<0.001) as well as CD146 MIF (89±136 vs. 36±95, p<0.001) and CD146 positivity rate [74/144 (51%) vs 25/98 (26%), p<0.001] when compared with Ph-neg ALLs (Table 4 and Table 5), confirming the key role played by the presence of CD146 in the peculiar phenotype of Ph+ B-ALL leukemic cells.
 |
|
 |
|
 |
|
No difference was evidenced in any of the groups in the expression of B-cell precursors classical marker such as CD19, CD22, CD20 and TdT.
To confirm the relevance of CD146 expression as an independent predictive factor for predicting the BCR::ABL1 rearrangement in B-ALLs. We carried out a multivariate study that included all the immunophenotypic variables considered important in Ph+ ALLs diagnosis. In such multivariate analysis (including CD146, CD10, CD13, CD20, CD33, CD66c, CD38) positivity of CD146 showed to be statistically associated with BCR::ABL1 rearrangement detection in B-ALLs [OR 1.01 (95% Cl: 1.01–1.02) (p=0.021)], along with CD13 [OR 1.02 (95% Cl: 1.01–1.04) (p<0.001)] and negative CD38 [OR 0.99 (95% Cl: 0.98–0.99) (p=0.005)].span style="font-family: 'Open Sans',sans-serif;">
Discussion
Nevertheless, a peculiar immunophenotypic pattern might be useful in case of limited access to molecular tests. Other studies have shown CD146 expression in B-ALLs and, in particular, in Ph+ ALLs. Nevertheless, in most of them, the sample size was small, or very heterogeneous groups were considered. The possible strength of our study relies on a homogeneous and relatively large cohort of newly diagnosed B-ALL cases referred to a single institution. CD146 expression as a useful prognostic biomarker has also been investigated by various reports and studies for different solid tumors.[21] In literature, reports about CD146 expression on progenitor cells in normal bone marrow or on hematological disease cells are limited. Cavazzini et al. found that the expression of CD146+cells was detected in 38.8% of B-cell ALL (14/38).[23] Interestingly, all the seven cases of Ph+ ALL were CD146-positive. They also found that the expression of CD13 and CD33 on adult B-ALL blasts was higher in the CD146-positive group, and CD146 expression was strongly associated with the presence of the Ph chromosome (p=0.001).[23] In a Chinese study, Xie et al. compared CD146 expression rates in adult and childhood B-ALL patients, which were 29.17% and 9.09%, respectively, showing a statistically significant difference (p< 0.05), probably due to higher incidence of Ph+ ALL in adult patients.[31] Another study involving 31 pediatric patients showed that for B-ALLs, the mean expression for CD146+ blasts was 51.347 ± 24.133, with the mean expression for CD146 on the blast cells was 51% ± 24%. Nevertheless, in this study, Zahran AM et al. could not correlate the percentage of CD146 expression with that of the Ph chromosome, possibly because of a shortage of data collection. Even so, CD146 was significantly associated with a lower response to induction therapy, suggesting a possible correlation with a subgroup of B-ALLs with an unfavorable prognosis.[32]
In summary, we can state that the evaluation in MFC panels of antigens like CD146, CD13, CD33, CD38, and CD66c, along with classical B-ALLs MFC markers (such as CD10, CD19, CD22, CD34, TdT), can aid the diagnosis of ALL and can partially rapidly suggest a distinction between molecularly negative ALL cases and Ph+ ALL cases.
In our cohort, Ph+ ALLs showed higher expression of CD146 in terms of mean percentage, positivity rate, and in term of MFI. In addition, myeloid markers such as CD33 and CD13, as already known,[26-27] are highly expressed in Ph+ ALLs and Ph-like ALLs, helping in the distinction from molecularly negative cases. Indeed, our data shows that CD13 and CD66c have higher expressions in Ph+ ALLs compared both with Ph-like ALLs and with molecularly negative ALLs. Hence, along with the expression of myeloid markers, CD146 expression is likely to represent an aberrant marker frequently associated with the t(9;22)(q34;q11)/BCR::ABL1, making possible to suspect a Ph+ ALL and prompting a “fast-track” for the detection of Ph chromosome. Indeed, Ph+ ALL blasts are characterized by the co-expression of myeloid markers CD13/CD33/CD66c with a low expression of CD38 and by the expression of CD146, as shown in the literature and in our data. On the other hand, this peculiar CD146 expression and MFI did not emerge in Ph-like ALLs. In addition, in our study, CD146 was never expressed in the entire blast population, so its role in minimal residual disease (MRD) remains limited. Further studies are necessary to establish CD146's role in the diagnosis, monitoring, and eventual relapse of ALLs.
Conclusions
Acknowledgments
Author Contributions
References
- Gökbuget N, Boissel N, Chiaretti S, Dombret H, Doubek M, Fielding AK, Foà R, Giebel S, Hoelzer D, Hunault M, Marks DI, Martinelli G, Ottmann O, Rijneveld AW, Rousselot P, Ribera JM, Bassan R; Management of ALL in Adults: 2023 ELN Recommendations from a European Expert Panel. Blood. 2024 Feb. https://doi.org/10.1182/blood.2023023568 PMid:38306595
- Hoelzer D, Bassan R, Dombret H, Fielding A, Ribera JM, Buske C; ESMO Guidelines Committee. Acute lymphoblastic leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016 Sep;27(suppl 5): v69-v82.https://doi.org/10.1093/annonc/mdw025 PMid:27056999
- Szczepański T, Van der Velden VH, Van Dongen JJ; Flow-cytometric Immunophenotyping of Normal and Malignant Lymphocytes. Clin Chem Lab Med. 2006 Jan;44(suppl 7): v775-v796. https://doi.org/10.1515/CCLM.2006.146 PMid:16776621
- S.R.,Krishnan S.,Bhatacharyya A.,Das A.,Kumar J.,Bhave S.,Radhakrishnan V., Nair R.,Chandy M.,Arora N.,Mishra D.Spectrum and immunophenotypic profile of acute leukemia: a tertiary center flow cytometry experience. Mediterr J Hematol Infect Dis 2019, 11(1): e2019017, https://doi.org/10.4084/mjhid.2019.017PMid:30858955 PMCid:PMC6402547
- Béné MC, Castoldi G, Knapp W, Ludwig WD, Matutes E, Orfao A, Van't Veer MB; Proposals for the Immunological Classification of Acute Leukemias. European Group for the Immunological Characterization of Leukemias (EGIL). Leukemia. 1995 Oct;9(suppl 10): v1783-v1786.http://europepmc.org/abstract/MED/7564526
- Béné MC, Nebe T, Bettelheim P, Buldini B, Bumbea H, Kern W, Lacombe F, Lemez P, Marinov I, Matutes E, Maynadié M, Oelschlagel U, Orfao A, Schabath R, Solenthaler M, Tschurtschenthaler G, Vladareanu AM, Zini G, Faure GC, Porwit A; Immunophenotyping of Acute Leukemia and Lymphoproliferative Disorders: A Consensus Proposal of the European LeukemiaNet Work Package 10. Leukemia. 2011 Apr;25(suppl 4): v567-v574.https://doi.org/10.1038/leu.2010.312 PMid:21252983
- Kulis J, Sędek Ł, Słota Ł, Perkowski B, Szczepański T; Commonly Assessed Markers in Childhood BCP-ALL Diagnostic Panels and Their Association with Genetic Aberrations and Outcome Prediction. Genes Basel 2022 Jul;13(suppl 8): v1374. https://doi.org/10.3390/genes13081374 PMid:36011285 PMCid:PMC9407579
- Nowell
PC, Hungerford DA; Chromosome Studies on Normal and Leukemic Human
Leukocytes. J Natl Cancer Inst 1960 Jul:
v85-v109.https://doi.org/10.1093/jnci/25.1.85
- Groffen J, Stephenson JR, Heisterkamp N, De Klein A, Bartram CR, Grosveld G; Philadelphia Chromosomal Breakpoints Are Clustered Within a Limited Region, Bcr, on Chromosome 22. Cell. 1984 Jan;36(suppl 1): v93-v99.https://doi.org/10.1016/0092-8674(84)90077-1 PMid:6319012
- Chiaretti S, Vitale A, Cazzaniga G, Orlando SM, Silvestri D, Fazi P, Valsecchi MG, Elia L, Testi AM, Mancini F, Conter V, Te Kronnie G, Ferrara F, Di Raimondo F, Tedeschi A, Fioritoni G, Fabbiano F, Meloni G, Specchia G, Pizzolo G, Mandelli F, Guarini A, Basso G, Biondi A, Foà R; Clinico-biological Features of 5202 Patients With Acute Lymphoblastic Leukemia Enrolled in the Italian AIEOP and GIMEMA Protocols and Stratified in Age Cohorts. Haematologica. 2013 Nov;98(suppl 11): v1702-v1710.https://doi.org/10.3324/haematol.2012.080432 PMid:23716539 PMCid:PMC3815170
- Mancini M, Scappaticci D, Cimino G, Nanni M, Derme V, Elia L, Tafuri A, Vignetti M, Vitale A, Cuneo A, Castoldi G, Saglio G, Pane F, Mecucci C, Camera A, Specchia G, Tedeschi A, Di Raimondo F, Fioritoni G, Fabbiano F, Marmont F, Ferrara F, Cascavilla N, Todeschini G, Nobile F, Kropp MG, Leoni P, Tabilio A, Luppi M, Annino L, Mandelli F, Foà R; A Comprehensive Genetic Classification of Adult Acute Lymphoblastic Leukemia (ALL): Analysis of the GIMEMA 0496 Protocol. Blood. 2005 May;105(suppl 9): v3434-v3441.https://doi.org/10.1182/blood-2004-07-2922
- Burmeister T, Schwartz S, Bartram CR, Gökbuget N, Hoelzer D, Thiel E; Patients' age and BCR-ABL frequency in adult B-precursor ALL: a retrospective analysis from the GMALL study group. Blood. 2008 Aug;112 (suppl 3): v918-v919.https://doi.org/10.1182/blood-2008-04-149286 PMid:18650471
- Foà R, Chiaretti S; Dasatinib-Blinatumomab for Ph-Positive ALL. Reply. N Engl J Med. 2021 Jan;384(suppl 4): v384.https://doi.org/10.1056/NEJMc2033785
- Foa' R, Bassan R, Elia L, Piciocchi A, Soddu S, Messina M, Ferrara F, Lunghi M, Mulè A, Bonifacio M, Fracchiolla N, Salutari P, Fazi P, Guarini A, Rambaldi A, Chiaretti S; Long-Term Results of the Dasatinib-Blinatumomab Protocol for Adult Philadelphia-Positive ALL. J Clin Oncol 2024 Mar;42(suppl 8): v881-v885.https://doi.org/10.1200/JCO.23.01075 PMid:38127722 PMCid:PMC10927329
- Den Boer ML, Van Slegtenhorst M, De Menezes RX, Cheok MH, Buijs-Gladdines JG, Peters ST, Van Zutven L, H Beverloo B, Van der Spek P J, Escherich G, Horstmann MA, Janka-Schaub GE, Kamps WA, Evans WE, Pieters R; A Subtype of Childhood Acute Lymphoblastic Leukaemia With Poor Treatment Outcome: A Genome-wide Classification Study. Lancet Oncol. 2009 Feb;10(suppl 2): v125-v134.https://doi.org/10.1016/S1470-2045(08)70339-5 PMid:19138562
- Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, Ma J, Liu W, Cheng C, Schulman BA, Harvey RC, Chen IM, Clifford RJ, Carroll WL, Reaman G, Bowman WP, Devidas M, Gerhard DS, Yang W, Relling MV, Shurtleff SA, Campana D, Borowitz MJ, Pui CH, Smith M, Hunger SP, Willman CL, Downing JR; Children's Oncology Group. Deletion of IKZF1 and Prognosis in Acute Lymphoblastic Leukemia. N Engl J Med. 2009 Jan;360(suppl 5): v470-v480.https://doi.org/10.1056/NEJMoa0808253 PMid:19129520 PMCid:PMC2674612
- Fazio F, Cunsolo G, Mancini F, De Propris DP, Piciocchi A, Arena V, Messina M, Ansuinelli M, Taherinasab AT, Apicella V, Vitale V, Chiaretti S, Guarini A, Del Giudice I, Foà R; Blast Morphology in the Diagnostic Workup of Ph-like Acute Lymphoblastic Leukemia. Leuk Lymphoma. 2022 Jun;63(suppl 6): v1512-v1514.https://doi.org/10.1080/10428194.2022.2032035 PMid:35133227
- Chiaretti S, Messina M, Foà R; BCR/ABL1-like Acute Lymphoblastic Leukemia: How to Diagnose and Treat? Cancer. 2019 Jan;125(suppl 2): v194-v204.https://doi.org/10.1002/cncr.31848 PMid:30561755
- Chiaretti S, Messina M, Della Starza I, Piciocchi A, Cafforio L, Cavalli M, Taherinasab A, Ansuinelli M, Elia L, Albertini Petroni G, La Starza R, Canichella M, Lauretti A, Puzzolo MC, Pierini V, Santoro A, Spinelli O, Apicella V, Capria S, Di Raimondo F, De Fabritiis P, Papayannidis C, Candoni A, Cairoli R, Cerrano M, Fracchiolla N, Mattei D, Cattaneo C, Vitale A, Crea E, Fazi P, Mecucci C, Rambaldi A, Guarini A, Bassan R, Foà R; Philadelphia-like Acute Lymphoblastic Leukemia Is Associated With Minimal Residual Disease Persistence and Poor Outcome. First Report of the Minimal Residual Disease-oriented GIMEMA LAL1913. Haematologica. 2021 Jun;106(suppl 6): v1559-v1568.https://doi.org/10.3324/haematol.2020.247973 PMid:32467145 PMCid:PMC8168510
- Lehmann JM, Riethmüller G, Johnson JP; MUC18, a Marker of Tumor Progression in Human Melanoma, Shows Sequence Similarity to the Neural Cell Adhesion Molecules of the Immunoglobulin Superfamily. Proc Natl Acad Sci U S A. 1989 Dec;86(suppl 24): v9891-v9895.https://doi.org/10.1073/pnas.86.24.9891 PMid:2602381 PMCid:PMC298608
- Ouhtit A, Gaur RL, Zakaria YA, Fernando A, Thouta R, Trappey AK, Abdraboh ME, El-Sayyad HI, Rao P, Raj MG; Towards Understanding the Mode of Action of the Multifaceted Cell Adhesion Receptor CD146. Biochim Biophys Acta. Reviews on Cancer. 2009 Apr;1795(suppl 2): v130-v136.https://doi.org/10.1016/j.bbcan.2009.01.002 PMid:19356677
- Zeng P, Li H, Lu PH, Zhou LN, Tang M, Liu CY, Chen MB; Prognostic Value of CD146 in Solid Tumor: A Systematic Review and Meta-analysis. Sci Rep. 2017 Jun;7(suppl 1): v4223.https://doi.org/10.1038/s41598-017-01061-3 PMid:28652617 PMCid:PMC5484668
- Cavazzini F, Campioni D, Ferrari L, Buldini B, Bardi MA, Michielotto B, Lazzari MC, Ongari M, Dabusti M, Daghia G, Sofritti O, Basso G, Lanza F, Cuneo A; Expression of the Immunoglobulin Superfamily Cell Membrane Adhesion Molecule CD146 in Acute Leukemia. Cytometry B Clin Cytom. 2016 May;90(suppl 3): v247-v256.https://doi.org/10.1002/cyto.b.21267 PMid:26102234
- Chiaretti S, Messina M, Grammatico S, Piciocchi A, Fedullo AL, Di Giacomo F, Peragine N, Gianfelici V, Lauretti A, Bareja R, Martelli MP, Vignetti M, Apicella V, Vitale A, Li LS, Salek C, Elemento O, Inghirami G, Weinstock DM, Guarini A, Foà R; Rapid Identification of BCR/ABL1‐like Acute Lymphoblastic Leukaemia Patients Using a Predictive Statistical Model Based on Quantitative Real Time‐polymerase Chain Reaction: Clinical, Prognostic and Therapeutic Implications. Br J Haematol. 2018 Jun;181(suppl 5): v642-v652.https://doi.org/10.1111/bjh.15251 PMid:29675955 PMCid:PMC5975184
- Alaggio R, Amador C, Anagnostopoulos I, Attygalle AD, Araujo IBO, Berti E, Bhagat G, Borges AM, Boyer D, Calaminici M, Chadburn A, Chan JKC, Cheuk W, Chng WJ, Choi JK, Chuang SS, Coupland SE, Czader M, Dave SS, de Jong D, Du MQ, Elenitoba-Johnson KS, Ferry J, Geyer J, Gratzinger D, Guitart J, Gujral S, Harris M, Harrison CJ, Hartmann S, Hochhaus A, Jansen PM, Karube K, Kempf W, Khoury J, Kimura H, Klapper W, Kovach AE, Kumar S, Lazar AJ, Lazzi S, Leoncini L, Leung N, Leventaki V, Li XQ, Lim MS, Liu WP, Louissaint A Jr, Marcogliese A, Medeiros LJ, Michal M, Miranda RN, Mitteldorf C, Montes-Moreno S, Morice W, Nardi V, Naresh KN, Natkunam Y, Ng SB, Oschlies I, Ott G, Parrens M, Pulitzer M, Rajkumar SV, Rawstron AC, Rech K, Rosenwald A, Said J, Sarkozy C, Sayed S, Saygin C, Schuh A, Sewell W, Siebert R, Sohani AR, Tooze R, Traverse-Glehen A, Vega F, Vergier B, Wechalekar AD, Wood B, Xerri L, Xiao W. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. 2022 Jul;36(suppl 7): v1720-v1748. Epub 2022 Jun 22. Erratum in: Leukemia. 2023 Sep;37(suppl 9): v1944-v1951.https://doi.org/10.1038/s41375-022-01620-2 PMid:35732829 PMCid:PMC9214472
- Theunissen P, Mejstrikova E, Sedek L, Van der Sluijs-Gelling AJ, Gaipa G, Bartels M, Sobral da Costa E, Kotrová M, Novakova M, Sonneveld E, Buracchi C, Bonaccorso P, Oliveira E , Te Marvelde JG, Szczepanski T, Lhermitte L, Hrusak O, Lecrevisse Q, Grigore GE, Froňková E, Trka J, Brüggemann M, Orfao A, Van Dongen JJ, Van der Velden VH; EuroFlow Consortium; Standardized Flow Cytometry for Highly Sensitive MRD Measurements in B-cell Acute Lymphoblastic Leukemia. Blood. 2017 Jan;129(suppl 3): v347-v357.https://doi.org/10.1182/blood-2016-07-726307 PMid:27903527 PMCid:PMC5291958
- Bisegna M.L., Peragine N., Elia L., Matarazzo M., Milani M.L., Intoppa S., Di Trani M., Malfona F., Martelli M., De Propris M.S.NG2 molecule expression in acute lymphoblastic leukemia B cells: a flow-cytometric marker for the rapid identification of KMT2A gene rearrangements.Mediterr J Hematol Infect Dis 2024, 16(1): e2024018https://doi.org/10.4084/MJHID.2024.018 PMid:38468826 PMCid:PMC10927233
- Elia L, Mancini M, Moleti L, Meloni G, Buffolino S, Krampera M, De Rossi G, Foà R, Cimino G; A Multiplex Reverse Transcriptase-polymerase Chain Reaction Strategy for the Diagnostic Molecular Screening of Chimeric Genes: A Clinical Evaluation on 170 Patients With Acute Lymphoblastic Leukemia. Haematologica. 2003 Mar;88(Suppl 3): v275-v279.https://pubmed.ncbi.nlm.nih.gov/12651265
- Tabernero MD, Bortoluci AM, Alaejos I, López-Berges MC, Rasillo A, García-Sanz R, García M, Sayagués JM, González M, Mateo G, San Miguel JF, Orfao A; Adult Precursor B-ALL With BCR/ABL Gene Rearrangements Displays a Unique Immunophenotype Based on the Pattern of CD10, CD34, CD13 and CD38 Expression. Leukemia. 2001 Mar;15(suppl 3): v406-v414.https://doi.org/10.1038/sj.leu.2402060 PMid:11237064
- Corrente F, Bellesi S, Metafuni E, Puggioni PL, Marietti S, Ciminello AM, Za T, Sorà F, Fianchi L, Sica S, De Stefano V, Chiusolo P; Role of Flow‐cytometric Immunophenotyping in Prediction of BCR/ABL1 Gene Rearrangement in Adult B‐cell Acute Lymphoblastic Leukemia. Cytometry B Clin Cytom. 2018 May;94(suppl 3): v468-v476.https://doi.org/10.1002/cyto.b.21605 PMid:29220871
- Xie XQ, Wang WM, Gan SL, Chen SM, Zhang QT, Xie XS, Liu YF, Cheng YD, Liu YF, Xue-Ju Xu 1, Sun H; Expression of CD146 in Adult and Children's Acute B Cell Lymphoblastic Leukemia and Its Significance. Journal of Experimental Hematology/Chinese Association of Pathophysiology. 2017 Febb;25(suppl 1): v30-v34.https://doi.org/10.7534/j.issn.1009-2137.2017.01.005
- Zahran AM, El-Badawy O, Elsayh KI, Mohamed WMY, Riad KF, Abdel-Rahim MH, Rayan A; Upregulation of CD146 in Pediatric B-Cell Acute Lymphocytic Leukemia and Its Implications on Treatment Outcomes. J Immunol Res. 2020 Feb; v1-v13.https://doi.org/10.1155/2020/9736159 PMid:32090132 PMCid:PMC7031726
Supplementary Files
 |
|
 |
|
 |
|

