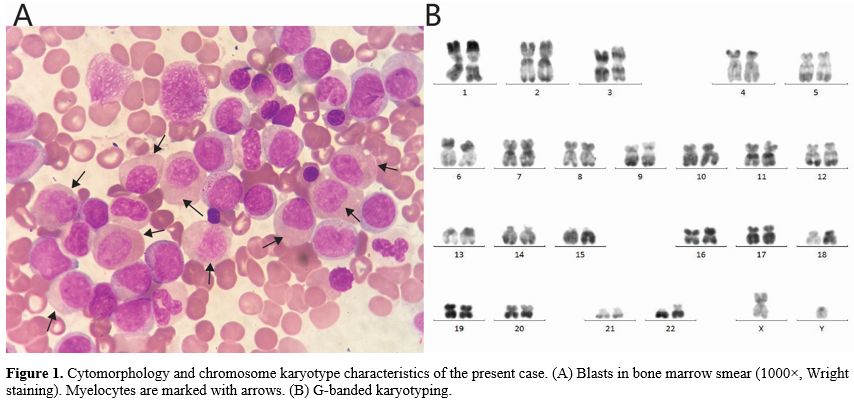
The patient was admitted to our hospital for the first time due to "leukocytosis found for 2 days". Routine blood tests indicated a white blood cell count of 23.08 × 10^9/L, neutrophil count of 9.65 × 10^9/L, monocyte count of 8.65 × 10^9/L, lymphocyte count of 4.74 × 10^9/L, hemoglobin level of 90 g/L, and platelet count of 19 × 10^9/L. Bone marrow smears showed that the proportions of myeloblasts, promyelocytes, abnormal neutrophils, and promonocytes were 4.5%, 2%, 28.5%, and 6%, respectively. Late erythroblasts with binucleated or petal-like nuclei were identified (Figure 1A). Blood smears revealed 15% myeloblasts, 12% myelocytes, 3% promonocytes, and 22% monocytes. Flow cytometry indicated that myeloblasts accounted for 21.54% of nuclear cells and expressed CD34, CD117, CD38, CD33, CD13, CD123, and HLA-DR. Promonocytes constituted 16.67% and exhibited the phenotypes CD38, CD64, CD123, CD33, CD13, CD36, and HLA-DR, with weak expression of CD11b. The results of chromosome karyotype analysis were normal (Figure 1B).
Genetic mutation testing revealed FLT3-ITD (5.2%), KRAS c.38G>A (6%), SRSF2 c.284C>A (47.8%), ASXL1 c.2309C>A (47.4%), RUNX1 c.941_942dup (47.4%), STAG2 c.1821+2T>C (94.7%), KMT2D c.15671C>A (47.9%), and TET2 c.2604T>G (47.6%). The diagnosis was acute myeloid leukemia (AML). The patient received a regimen of azacitidine (140 mg d1-7) and venetoclax (100 mg d1, 200 mg d2, 400 mg d3-14) for 2 courses, none of which achieved remission. The patient was eventually lost to follow-up.
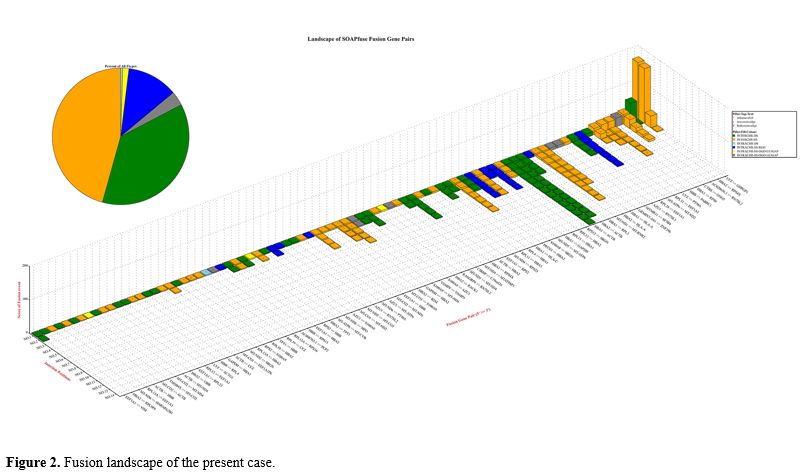
Total RNA was extracted from the bone marrow mononuclear cells. Transcriptome sequencing was performed using the Illumina HiSeq 2500 instrument (Illumina, San Diego, CA). Employing SOAPfuse software, a series of fusion events was predicted (Figure 2).
In the 1950s, Yang and Yan et al. in China defined a unique form of AML. The predominant leukemic cells in the bone marrow are abnormal myelocytes. Compared with other AML leukemic cells, these abnormal myelocytes are larger, exhibit a relatively lower nuclear-to-cytoplasmic ratio, and show asynchronous development of the nucleus and cytoplasm. The nucleus contains fine chromatin dotted with one or two nucleoli. The abundant cytoplasm is typically basophilic. In the invagination of the nucleus, a homogeneous salmon coloration can be observed. The majority of these cells express stem/progenitor cell markers. Most reports suggest a high expression of CD34 and HLA-DR. CD15 is relatively highly expressed, whereas CD33 and CD13 are expressed at low levels, and CD7 is rarely expressed. The results of the literature review indicate that 40-98.2% of M2b blasts express CD19, and 20-71.4% express CD56. According to China's revised FAB classification of leukemia, this form is classified as the M2b subtype. Most cases of M2b harbor the RUNX1::RUNX1T1 fusion gene and the t(8;21)(q22;q22.1) translocation. Morphological features of M2b are also observed in other myeloid tumors, such as the acute phase of chronic myeloid leukemia, myelodysplastic syndromes, M4, and M6. However, the RUNX1::RUNX1T1 fusion gene is absent in all of these cases.[1-3]
In the case we present, cell morphology demonstrated a predominance of abnormal myelocytes (28.5%), while typical myeloblasts were rare (4.5%). The percentage of myeloblasts detected by flow cytometry was 21.54%, and these cells expressed stem/progenitor cell markers. Interestingly, the monocyte subpopulation exhibited similar characteristics, with cell morphology showing more mature features compared to the results from flow cytometry.
RNA sequencing (RNA-seq) revealed a series of fusion events, notably the absence of RUNX1::RUNX1T1 and other frequent recurrent fusion genes. There are significant advantages of interchromosomal fusion compared with intrachromosomal fusion, as both coding and non-coding genes are involved. Some known oncogenic genes, such as ZNF296 and SF3B4, can be identified as fusion partners.[4,5] These partner genes are involved in various aspects of cellular function, including the regulation of transcription, pre-mRNA splicing, mRNA stabilization, tRNA delivery, protein synthesis, and degradation, the insertion of secretory proteins, glucose metabolic processes, carbohydrate metabolism, ribosome construction, enzyme activity facilitation, inflammatory mediation, cell motility, cytoskeleton maintenance, apoptosis, and cell division. Multiple fusion genes are common in tumors and have different potential contributions to cancer development.[6]
In summary, we report a case of AML characterized by abnormal myelocyte and monocyte morphology in which the fusion gene landscape was delineated. Further cases are needed to elucidate the genomic features associated with this subtype.
References
- Yushu H, Shougeng B, Zhijian X, et al. (1999) Acute myeloid leukemia M2b. Haematologica 84:193-194
- Xiao Z, Hao Y, Bian S (1997) Acute myeloid leukemia M2B (subacute myeloid leukemia) in China. Leukemia Res 21:351-352. https://doi.org/10.1016/S0145-2126(96)00101-4 PMid:9150353
- 杨崇礼, 张新伟, 肖志坚, et al (2004) 急性髓系白血病M2b的研究进展. 白血病.淋巴瘤 13:9
- Mizoue
Y, Ikeda T, Ikegami T, et al. (2023) The stem cell transcription factor
ZFP296 transforms NIH3T3 cells and promotes anchorage-independent
growth of cancer cells. Int J Dev Biol 67:147-153. https://doi.org/10.1387/ijdb.230143hk PMid:38334180
- Kidogami
S, Iguchi T, Sato K, et al. (2020) SF3B4 Plays an Oncogenic Role in
Esophageal Squamous Cell Carcinoma. Anticancer Res 40:2941-2946. https://doi.org/10.21873/anticanres.14272 PMid:32366446
- Stephens
PJ, McBride DJ, Lin M-L, et al. (2009) Complex landscapes of somatic
rearrangement in human breast cancer genomes. Nature 462:1005-1010. https://doi.org/10.1038/nature08645 PMid:20033038 PMCid:PMC3398135