THE IMPORTANCE OF EPIGENETIC ALTERATIONS IN THE DEVELOPMENT OF EPSTEIN-BARR VIRUS-RELATED LYMPHOMAS
Main Article Content
Keywords
carcinogenesis, DNA methylation, patho-epigenetics, epigenetic reprogramming, Epstein-Barr virus, epigenetic dysregulation
Abstract
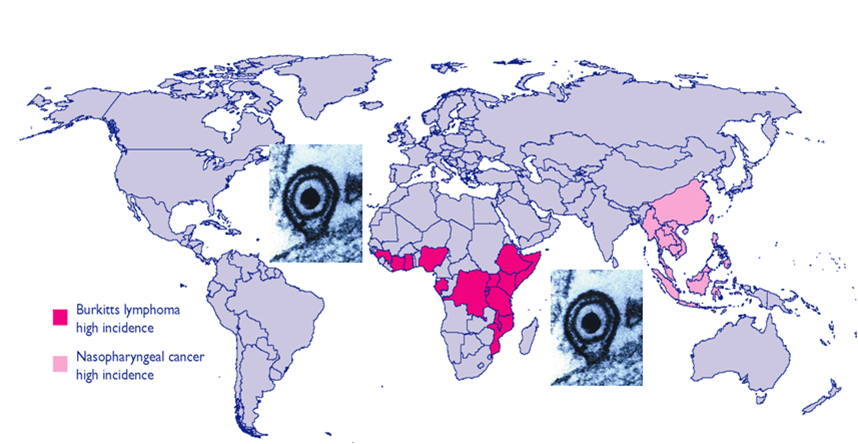
Epstein-Barr virus (EBV), a human gammaherpesvirus, is associated with a series of malignant tumors. These include lymphomas (Burkitt’s lymphoma, Hodgkin’s disease, T/NK-cell lymphoma, post-transplant lymphoproliferative disease, AIDS-associated lymphoma, X-linked lymphoproliferative syndrome), carcinomas (nasopharyngeal carcinoma, gastric carcinoma, carcinomas of major salivary glands, thymic carcinoma, mammary carcinoma) and a sarcoma (leiomyosarcoma). The latent EBV genomes persist in the tumor cells as circular episomes, co-replicating with the cellular DNA once per cell cycle. The expression of latent EBV genes is cell type specific due to the strict epigenetic control of their promoters. DNA methylation, histone modifications and binding of key cellular regulatory proteins contribute to the regulation of alternative promoters for transcripts encoding the nuclear antigens EBNA1 to 6 and affect the activity of promoters for transcripts encoding transmembrane proteins (LMP1, LMP2A, LMP2B). In addition to genes transcribed by RNA polymerase II, there are also two RNA polymerase III transcribed genes in the EBV genome (EBER 1 and 2). The 5’ and internal regulatory sequences of EBER 1 and 2 transcription units are invariably unmethylated. The highly abundant EBER 1 and 2 RNAs are not translated to protein. Based on the cell type specific epigenetic marks associated with latent EBV genomes one can distinguish between viral epigenotypes that differ in transcriptional activity in spite of having an identical (or nearly identical) DNA sequence. Whereas latent EBV genomes are regularly targeted by epigenetic control mechanisms in different cell types, EBV encoded proteins may, in turn, affect the activity of a set of cellular promoters by interacting with the very same epigenetic regulatory machinery. There are EBNA1 binding sites in the human genome. Because high affinity binding of EBNA1 to its recognition sites is known to specify sites of DNA demethylation, we suggest that binding of EBNA1 to its cellular target sites may elicit local demethylation and contribute thereby to the activation of silent cellular promoters. EBNA2 interacts with histone acetyltransferases, and EBNALP (EBNA5) coactivates transcription by displacing histone deacetylase 4 from EBNA2-bound promoter sites. EBNA3C (EBNA6) seems to be associated both with histone acetylases and deacetylases, although in separate complexes. LMP1, a transmembrane protein involved in malignant transformation, can affect both alternative systems of epigenetic memory, DNA methylation and the Polycomb-trithorax group of protein complexes. In epithelial cells LMP1 can up-regulate DNA methyltransferases and, in Hodgkin lymphoma cells, induce the Polycomb group protein Bmi-1. In addition, LMP1 can also modulate cellular gene expression programs by affecting, via the NF-?B pathway, levels of cellular microRNAs miR-146a and miR-155. These interactions may result in epigenetic dysregulation and subsequent cellular dysfunctions that may manifest in or contribute to the development of pathological changes (e.g. initiation and progression of malignant neoplasms; autoimmune phenomena; immunodeficiency). Thus, Epstein-Barr virus, similarly to other viruses and certain bacteria, may induce pathological changes by epigenetic reprogramming of host cells. Elucidation of the epigenetic consequences of EBV-host interactions (within the framework of the emerging new field of patho-epigenetics) may have important implications for therapy and disease prevention, because epigenetic processes are reversible and continuous silencing of EBV genes contributing to patho-epigenetic changes may prevent disease development.
Downloads
Abstract 741
PDF Downloads 398
HTML Downloads 2811